Difference between revisions of "SsuA"
(→Expression and regulation) |
|||
| Line 90: | Line 90: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ssuB]]-[[ssuA]]-[[ssuC]]-[[ssuD]]-[[ygaN]]'' | + | * '''Operon:''' ''[[ssuB]]-[[ssuA]]-[[ssuC]]-[[ssuD]]-[[ygaN]]'' {{PubMed|9782504}} |
* '''[[Sigma factor]]:''' [[SigA]] | * '''[[Sigma factor]]:''' [[SigA]] | ||
| − | * '''Regulation:''' | + | * '''Regulation:''' |
** repressed in the presence of cysteine ([[CymR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/16513748 PubMed] | ** repressed in the presence of cysteine ([[CymR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/16513748 PubMed] | ||
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' |
** [[CymR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/16513748 PubMed] | ** [[CymR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/16513748 PubMed] | ||
Revision as of 19:25, 2 September 2009
- Description: aliphatic sulfonate ABC transporter (binding lipoprotein)
| Gene name | ssuA |
| Synonyms | ygbA, yzeA |
| Essential | no |
| Product | aliphatic sulfonate ABC transporter (binding lipoprotein) |
| Function | sulfonate uptake |
| Metabolic function and regulation of this protein in SubtiPathways: Cys, Met & Sulfate assimilation | |
| MW, pI | 36 kDa, 4.85 |
| Gene length, protein length | 996 bp, 332 aa |
| Immediate neighbours | ssuB, ssuC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 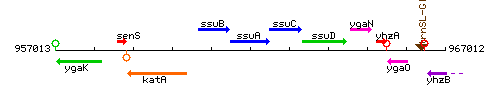
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08840
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial solute-binding protein ssuA/tauA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P40400
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Sergine Even, Pierre Burguière, Sandrine Auger, Olga Soutourina, Antoine Danchin, Isabelle Martin-Verstraete
Global control of cysteine metabolism by CymR in Bacillus subtilis.
J Bacteriol: 2006, 188(6);2184-97
[PubMed:16513748]
[WorldCat.org]
[DOI]
(P p)
Kyle N Erwin, Shunji Nakano, Peter Zuber
Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx.
J Bacteriol: 2005, 187(12);4042-9
[PubMed:15937167]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)
Jan R van der Ploeg, Nicola J Cummings, Thomas Leisinger, Ian F Connerton
Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates.
Microbiology (Reading): 1998, 144 ( Pt 9);2555-2561
[PubMed:9782504]
[WorldCat.org]
[DOI]
(P p)