Difference between revisions of "SrfAB"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 1: | Line 1: | ||
| − | + | * '''Description:''' surfactin synthetase / competence <br/><br/> | |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || antibiotic synthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || antibiotic synthesis | ||
| − | |||
| − | |||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 400 kDa, 4.903 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 400 kDa, 4.903 | ||
Revision as of 11:05, 7 August 2012
- Description: surfactin synthetase / competence
| Gene name | srfAB |
| Synonyms | comL |
| Essential | no |
| Product | surfactin synthetase / competence |
| Function | antibiotic synthesis |
| MW, pI | 400 kDa, 4.903 |
| Gene length, protein length | 10761 bp, 3587 aa |
| Immediate neighbours | srfAA, comS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed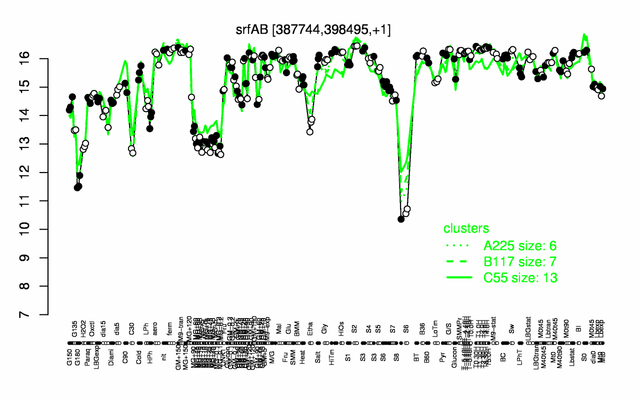
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds, membrane proteins, phosphoproteins
This gene is a member of the following regulons
Abh regulon, CodY regulon, ComA regulon, PerR regulon, Spx regulon
The gene
Basic information
- Locus tag: BSU03490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ATP-dependent AMP-binding enzyme family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane PubMed
Database entries
- Structure:
- UniProt: Q04747
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Massimiliano Marvasi, Pieter T Visscher, Lilliam Casillas Martinez
Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis.
FEMS Microbiol Lett: 2010, 313(1);1-9
[PubMed:20735481]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Yasutaro Fujita
Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression.
FEMS Microbiol Lett: 2007, 268(1);73-80
[PubMed:17227471]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Paul D Straight, Michael A Fischbach, Christopher T Walsh, David Z Rudner, Roberto Kolter
A singular enzymatic megacomplex from Bacillus subtilis.
Proc Natl Acad Sci U S A: 2007, 104(1);305-10
[PubMed:17190806]
[WorldCat.org]
[DOI]
(P p)
Kentaro Hayashi, Taku Ohsawa, Kazuo Kobayashi, Naotake Ogasawara, Mitsuo Ogura
The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis.
J Bacteriol: 2005, 187(19);6659-67
[PubMed:16166527]
[WorldCat.org]
[DOI]
(P p)
Natalia Comella, Alan D Grossman
Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis.
Mol Microbiol: 2005, 57(4);1159-74
[PubMed:16091051]
[WorldCat.org]
[DOI]
(P p)
Shunji Nakano, Michiko M Nakano, Ying Zhang, Montira Leelakriangsak, Peter Zuber
A regulatory protein that interferes with activator-stimulated transcription in bacteria.
Proc Natl Acad Sci U S A: 2003, 100(7);4233-8
[PubMed:12642660]
[WorldCat.org]
[DOI]
(P p)
M S Turner, J D Helmann
Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis.
J Bacteriol: 2000, 182(18);5202-10
[PubMed:10960106]
[WorldCat.org]
[DOI]
(P p)
P Serror, A L Sonenshein
CodY is required for nutritional repression of Bacillus subtilis genetic competence.
J Bacteriol: 1996, 178(20);5910-5
[PubMed:8830686]
[WorldCat.org]
[DOI]
(P p)
D Vollenbroich, N Mehta, P Zuber, J Vater, R M Kamp
Analysis of surfactin synthetase subunits in srfA mutants of Bacillus subtilis OKB105.
J Bacteriol: 1994, 176(2);395-400
[PubMed:8288534]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, L A Xia, P Zuber
Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis.
J Bacteriol: 1991, 173(17);5487-93
[PubMed:1715856]
[WorldCat.org]
[DOI]
(P p)