Difference between revisions of "SpsL"
| Line 42: | Line 42: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[GerE regulon]]}}, | ||
{{SubtiWiki regulon|[[SigK regulon]]}} | {{SubtiWiki regulon|[[SigK regulon]]}} | ||
| Line 60: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 111: | Line 109: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** expressed late during sporulation in the mother cell ([[SigK]]) {{PubMed|15383836}} | + | ** expressed late during sporulation in the mother cell ([[SigK]], [[GerE]]) {{PubMed|25239894,15383836}} |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[GerE]]: transcription activation {{PubMed|25239894,15383836}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 136: | Line 135: | ||
=References= | =References= | ||
| − | <pubmed>9353933 15383836 22960854 </pubmed> | + | <pubmed>9353933 15383836 22960854 25239894</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:03, 23 September 2014
- Description: dTDP -4-dehydrorhamnose-3,5-epimerase, spore coat polysaccharide synthesis
| Gene name | spsL |
| Synonyms | ipa-73d |
| Essential | no |
| Product | dTDP -4-dehydrorhamnose-3,5-epimerase |
| Function | rhamnose biosynthesis, spore coat polysaccharide synthesis |
| Gene expression levels in SubtiExpress: spsL | |
| MW, pI | 17 kDa, 4.878 |
| Gene length, protein length | 453 bp, 151 aa |
| Immediate neighbours | sivA, spsK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 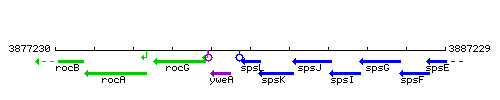
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed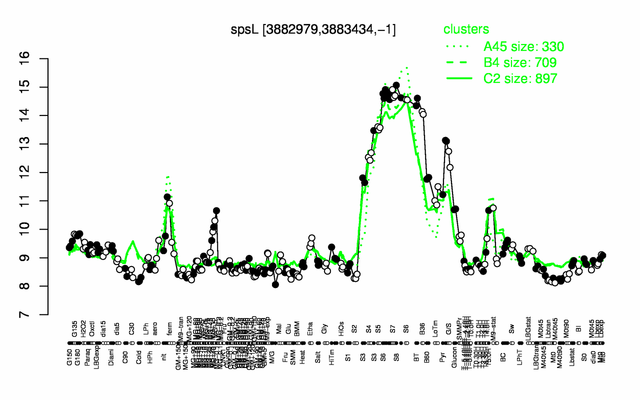
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37810
Phenotypes of a mutant
Database entries
- BsubCyc: BSU37810
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU37810
- Structure: 3RYK (dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) from Bacillus anthracis str. Ames with TDP and PPi bound, 32% identity, 56% similarity)
- UniProt: Q7WY56
- KEGG entry: [3]
- E.C. number: 5.1.3.13
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Giuseppina Cangiano, Teja Sirec, Cristina Panarella, Rachele Isticato, Loredana Baccigalupi, Maurilio De Felice, Ezio Ricca
The sps Gene Products Affect the Germination, Hydrophobicity, and Protein Adsorption of Bacillus subtilis Spores.
Appl Environ Microbiol: 2014, 80(23);7293-302
[PubMed:25239894]
[WorldCat.org]
[DOI]
(I p)
Germán Plata, Tobias Fuhrer, Tzu-Lin Hsiao, Uwe Sauer, Dennis Vitkup
Global probabilistic annotation of metabolic networks enables enzyme discovery.
Nat Chem Biol: 2012, 8(10);848-54
[PubMed:22960854]
[WorldCat.org]
[DOI]
(I p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)