Difference between revisions of "SpsI"
| Line 1: | Line 1: | ||
| − | * '''Description:''' spore coat polysaccharide synthesis <br/><br/> | + | * '''Description:''' glucose-1-phosphate thymidylyltransferase, spore coat polysaccharide synthesis <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || glucose-1-phosphate thymidylyltransferase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || spore coat polysaccharide synthesis | + | |style="background:#ABCDEF;" align="center"|'''Function''' || rhamnose biosynthesis, spore coat polysaccharide synthesis |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU37840 spsI] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU37840 spsI] | ||
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[miscellaneous metabolic pathways]]}}, | ||
{{SubtiWiki category|[[sporulation proteins]]}} | {{SubtiWiki category|[[sporulation proteins]]}} | ||
| Line 69: | Line 66: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** alpha-D-glucose-1-phosphate + dTTP --> dTDP-glucose {{PubMed|22960854}} | ||
* '''Protein family:''' glucose-1-phosphate thymidylyltransferase family (according to Swiss-Prot) | * '''Protein family:''' glucose-1-phosphate thymidylyltransferase family (according to Swiss-Prot) | ||
| Line 98: | Line 96: | ||
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU37840] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU37840] | ||
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.7.24 2.7.7.24] |
=== Additional information=== | === Additional information=== | ||
Revision as of 16:03, 18 September 2012
- Description: glucose-1-phosphate thymidylyltransferase, spore coat polysaccharide synthesis
| Gene name | spsI |
| Synonyms | ipa-71d |
| Essential | no |
| Product | glucose-1-phosphate thymidylyltransferase |
| Function | rhamnose biosynthesis, spore coat polysaccharide synthesis |
| Gene expression levels in SubtiExpress: spsI | |
| MW, pI | 27 kDa, 5.385 |
| Gene length, protein length | 738 bp, 246 aa |
| Immediate neighbours | spsJ, spsG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 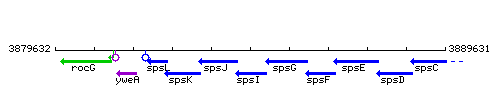
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed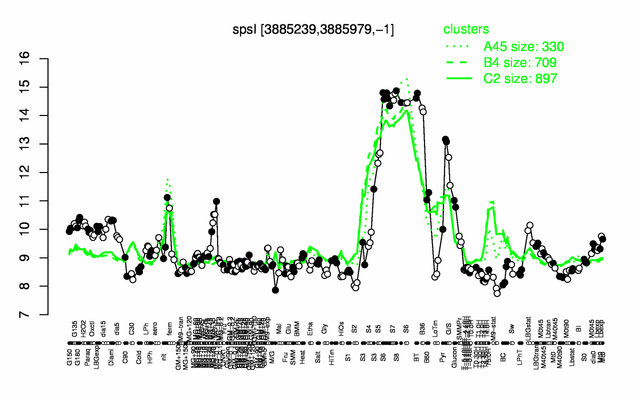
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37840
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- alpha-D-glucose-1-phosphate + dTTP --> dTDP-glucose PubMed
- Protein family: glucose-1-phosphate thymidylyltransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39629
- KEGG entry: [3]
- E.C. number: 2.7.7.24
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Germán Plata, Tobias Fuhrer, Tzu-Lin Hsiao, Uwe Sauer, Dennis Vitkup
Global probabilistic annotation of metabolic networks enables enzyme discovery.
Nat Chem Biol: 2012, 8(10);848-54
[PubMed:22960854]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)