Difference between revisions of "SpoVS"
(→Database entries) |
|||
| Line 53: | Line 53: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU16980&redirect=T BSU16980] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/spoVS.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/spoVS.html] | ||
| Line 87: | Line 88: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU16980&redirect=T BSU16980] | ||
* '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=2ek0 2EK0] (from ''Thermus thermophilus'', 56% identity) | * '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=2ek0 2EK0] (from ''Thermus thermophilus'', 56% identity) | ||
Revision as of 13:47, 2 April 2014
- Description: required for dehydratation of the spore core and assembly of the coat, mutation increases SigD-dependent gene expression, might act via SinR
| Gene name | spoVS |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | spore coat assembly, spore core dehydratation |
| Gene expression levels in SubtiExpress: spoVS | |
| MW, pI | 8 kDa, 6.475 |
| Gene length, protein length | 258 bp, 86 aa |
| Immediate neighbours | ymdB, tdh |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed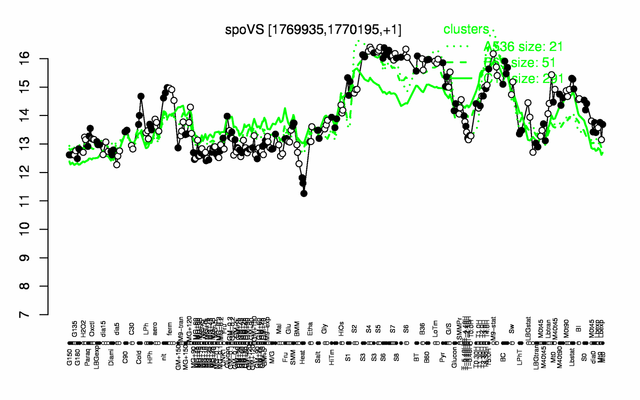
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16980
Phenotypes of a mutant
increased SigD-dependent gene expression, this might occur via SinR PubMed
Database entries
- BsubCyc: BSU16980
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU16980
- Structure: 2EK0 (from Thermus thermophilus, 56% identity)
- UniProt: P45693
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant: GP1601 (cat), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Daniel J Rigden, Michael Y Galperin
Sequence analysis of GerM and SpoVS, uncharacterized bacterial 'sporulation' proteins with widespread phylogenetic distribution.
Bioinformatics: 2008, 24(16);1793-7
[PubMed:18562273]
[WorldCat.org]
[DOI]
(I p)
Ana R Perez, Angelica Abanes-De Mello, Kit Pogliano
Suppression of engulfment defects in bacillus subtilis by elevated expression of the motility regulon.
J Bacteriol: 2006, 188(3);1159-64
[PubMed:16428420]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, A Driks, R Losick
Identification and characterization of sporulation gene spoVS from Bacillus subtilis.
J Bacteriol: 1995, 177(19);5628-35
[PubMed:7559352]
[WorldCat.org]
[DOI]
(P p)