Difference between revisions of "SodA"
| Line 41: | Line 41: | ||
{{SubtiWiki category|[[resistance against oxidative and electrophile stress]]}}, | {{SubtiWiki category|[[resistance against oxidative and electrophile stress]]}}, | ||
{{SubtiWiki category|[[membrane proteins]]}}, | {{SubtiWiki category|[[membrane proteins]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 61: | Line 62: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 79: | Line 77: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylation on Thr-34 AND Thr-70 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | * '''Modification:''' phosphorylation on Thr-34 AND Thr-70 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' manganese |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 119: | Line 117: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** The mRNA has a long 5' leader region. This may indicate RNA-based regulation {{PubMed|20525796}} | ** The mRNA has a long 5' leader region. This may indicate RNA-based regulation {{PubMed|20525796}} | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 140: | Line 139: | ||
=References= | =References= | ||
| − | <pubmed>9393707,9573176, 20525796,15805528 17218307, 9658017 10074093 22174384 22582280</pubmed> | + | <pubmed>9393707,9573176, 20525796,15805528 17218307, 9658017 10074093 22174384 22582280 15378759</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:21, 5 March 2014
- Description: superoxide dismutase, general stress protein, important for survival of ethanol and paraquat stresses and at low temperatures
| Gene name | sodA |
| Synonyms | yqgD |
| Essential | no |
| Product | superoxide dismutase |
| Function | detoxification of oxygen radicals |
| Gene expression levels in SubtiExpress: sodA | |
| MW, pI | 22 kDa, 5.203 |
| Gene length, protein length | 606 bp, 202 aa |
| Immediate neighbours | yqgE, yqgC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 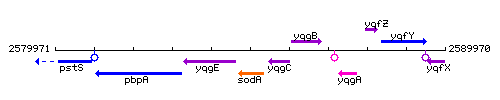
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed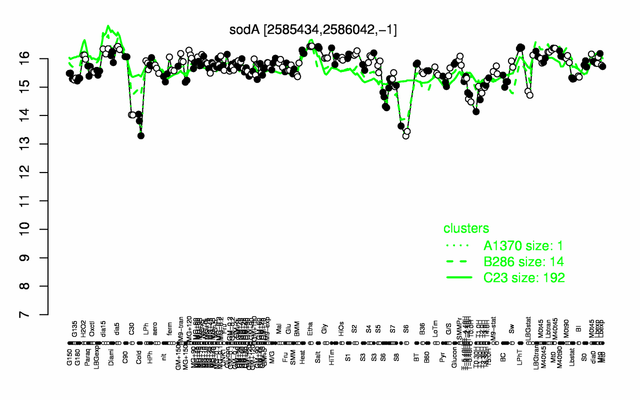
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress, membrane proteins, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25020
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 superoxide + 2 H+ = O2 + H2O2 (according to Swiss-Prot)
- Protein family: iron/manganese superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s): SodF
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Thr-34 AND Thr-70 PubMed
- Cofactors: manganese
- Effectors of protein activity:
- Localization: cytoplasm PubMed
Database entries
- Structure: 2RCV
- UniProt: P54375
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alexander Reder, Dirk Höper, Ulf Gerth, Michael Hecker
Contributions of individual σB-dependent general stress genes to oxidative stress resistance of Bacillus subtilis.
J Bacteriol: 2012, 194(14);3601-10
[PubMed:22582280]
[WorldCat.org]
[DOI]
(I p)
Wang Yung Tu, Susanne Pohl, Pijug Summpunn, Silvio Hering, Sandra Kerstan, Colin R Harwood
Comparative analysis of the responses of related pathogenic and environmental bacteria to oxidative stress.
Microbiology (Reading): 2012, 158(Pt 3);636-647
[PubMed:22174384]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
T Inaoka, Y Matsumura, T Tsuchido
SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis.
J Bacteriol: 1999, 181(6);1939-43
[PubMed:10074093]
[WorldCat.org]
[DOI]
(P p)
T Inaoka, Y Matsumura, T Tsuchido
Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis.
J Bacteriol: 1998, 180(14);3697-703
[PubMed:9658017]
[WorldCat.org]
[DOI]
(P p)
A O Henriques, L R Melsen, C P Moran
Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis.
J Bacteriol: 1998, 180(9);2285-91
[PubMed:9573176]
[WorldCat.org]
[DOI]
(P p)
L Casillas-Martinez, P Setlow
Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents.
J Bacteriol: 1997, 179(23);7420-5
[PubMed:9393707]
[WorldCat.org]
[DOI]
(P p)