Difference between revisions of "SodA"
| Line 78: | Line 78: | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2RCV 2RCV] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2RCV 2RCV] | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/P54375 P54375] |
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU25020] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU25020] | ||
Revision as of 13:37, 20 July 2009
- Description: superoxide dismutase, general stress protein
| Gene name | sodA |
| Synonyms | yqgD |
| Essential | no |
| Product | superoxide dismutase |
| Function | detoxification of oxygen radicals |
| MW, pI | 22 kDa, 5.203 |
| Gene length, protein length | 606 bp, 202 aa |
| Immediate neighbours | yqgE, yqgC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 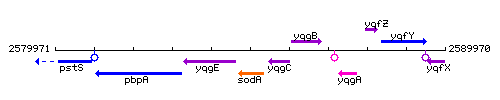
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25020
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 superoxide + 2 H+ = O2 + H2O2 (according to Swiss-Prot)
- Protein family: iron/manganese superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s): SodF
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Thr-34 AND Thr-70 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure: 2RCV
- UniProt: P54375
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
A O Henriques, L R Melsen, C P Moran
Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis.
J Bacteriol: 1998, 180(9);2285-91
[PubMed:9573176]
[WorldCat.org]
[DOI]
(P p)
L Casillas-Martinez, P Setlow
Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents.
J Bacteriol: 1997, 179(23);7420-5
[PubMed:9393707]
[WorldCat.org]
[DOI]
(P p)