Difference between revisions of "SnaB"
(→Basic information/ Evolution) |
(→Expression and regulation) |
||
| Line 104: | Line 104: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[yxeK]]-[[ | + | * '''Operon:''' ''[[yxeK]]-[[snaB]]-[[yxeM]]-[[yxeN]]-[[yxeO]]-[[sndB]]-[[yxeQ]]'' {{PubMed|10746760}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yxeL_4060307_4060804_-1 yxeL] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yxeL_4060307_4060804_-1 yxeL] {{PubMed|22383849}} | ||
Revision as of 11:48, 18 August 2013
- Description: sulphur N-acetylase, may be involved in the utilization of modified amino acids
| Gene name | snaB |
| Synonyms | yxeL |
| Essential | no |
| Product | sulphur N-acetylase |
| Function | unknown |
| Gene expression levels in SubtiExpress: snaB | |
| MW, pI | 18 kDa, 9.125 |
| Gene length, protein length | 495 bp, 165 aa |
| Immediate neighbours | yxeM, yxeK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 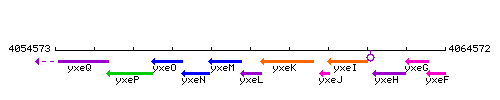
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed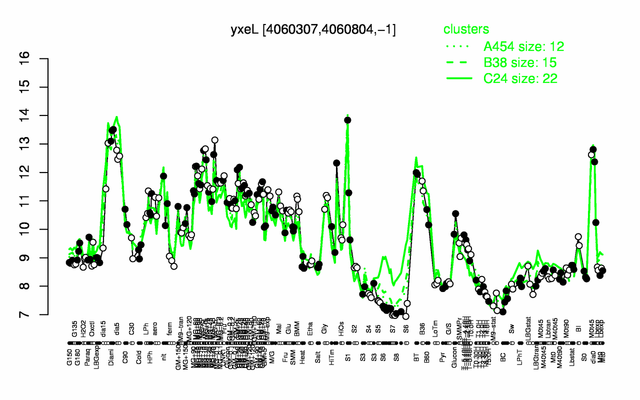
| |
Contents
Categories containing this gene/protein
poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39510
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: acetyltransferase family (according to Swiss-Prot)
- Paralogous protein(s): SnaA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54951
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Che-Man Chan, Antoine Danchin, Philippe Marlière, Agnieszka Sekowska
Paralogous metabolism: S-alkyl-cysteine degradation in Bacillus subtilis.
Environ Microbiol: 2014, 16(1);101-17
[PubMed:23944997]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Sergine Even, Pierre Burguière, Sandrine Auger, Olga Soutourina, Antoine Danchin, Isabelle Martin-Verstraete
Global control of cysteine metabolism by CymR in Bacillus subtilis.
J Bacteriol: 2006, 188(6);2184-97
[PubMed:16513748]
[WorldCat.org]
[DOI]
(P p)
Kyle N Erwin, Shunji Nakano, Peter Zuber
Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx.
J Bacteriol: 2005, 187(12);4042-9
[PubMed:15937167]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)