Difference between revisions of "SkfD"
(→Extended information on the protein) |
|||
| (32 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' This gene has been deleted due to its merger with ''[[skfC]]'' according to Barbe et al. (2009) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''skfD'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ybcT'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || unknown |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 34 kDa, 7.198 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 34 kDa, 7.198 | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || | ||
| − | |||
| − | |||
| − | |||
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ybcT_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ybcT_context.gif]] | ||
| + | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
|} | |} | ||
| Line 35: | Line 33: | ||
=== Basic information === | === Basic information === | ||
| − | * ''' | + | * '''Locus tag:''' BSU01940 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 41: | Line 39: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/skf.html] |
* '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12717] | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12717] | ||
| Line 70: | Line 68: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' membrane (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 78: | Line 76: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/O31426 O31426] |
| − | * '''KEGG entry:''' | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU01940] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 86: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[skfA]]-[[skfB]]-[[skfC]]-[[skfE]]-[[skfF]]-[[skfG]]-[[skfH]]'' [http://www.ncbi.nlm.nih.gov/pubmed/16816204 PubMed] |
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/pubmed/16452424 PubMed] |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** expressed under conditions that trigger sporulation ([[Spo0A]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647,15687200 PubMed] | ||
| + | ** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/15687200,17720793 PubMed] | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[Spo0A]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647,15687200 PubMed] | ||
| + | ** [[AbrB]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/15687200,17720793 PubMed] | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' Northern blotting during during phosphate limitation showed an intense 0.25 kb ''[[skfA]]''-specific transcript, and a weaker 6.5 kb ''[[skfA]]-[[skfB]]-[[skfC]]-[[skfE]]-[[skfF]]-[[skfG]]-[[skfH]]'' transcript. [http://www.ncbi.nlm.nih.gov/pubmed/16816204 PubMed] |
=Biological materials = | =Biological materials = | ||
| Line 117: | Line 119: | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed>20955377 </pubmed> | ||
| + | ==Original Publications== | ||
| + | <pubmed>12817086,15812018,,16816204,14651647,15687200,15687200,17720793,17449691,18840696 </pubmed> | ||
| − | + | [[Category:Protein-coding genes]] | |
Latest revision as of 20:48, 19 December 2011
- Description: This gene has been deleted due to its merger with skfC according to Barbe et al. (2009)
| Gene name | skfD |
| Synonyms | ybcT |
| Essential | no |
| Product | unknown |
| Function | |
| MW, pI | 34 kDa, 7.198 |
| Gene length, protein length | |
| Immediate neighbours | |
Genetic context 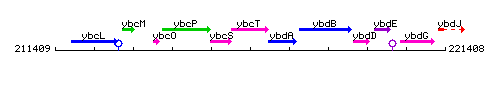
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU01940
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O31426
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information: Northern blotting during during phosphate limitation showed an intense 0.25 kb skfA-specific transcript, and a weaker 6.5 kb skfA-skfB-skfC-skfE-skfF-skfG-skfH transcript. PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
José Eduardo González-Pastor
Cannibalism: a social behavior in sporulating Bacillus subtilis.
FEMS Microbiol Rev: 2011, 35(3);415-24
[PubMed:20955377]
[WorldCat.org]
[DOI]
(I p)
Original Publications
Allison V Banse, Arnaud Chastanet, Lilah Rahn-Lee, Errett C Hobbs, Richard Losick
Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2008, 105(40);15547-52
[PubMed:18840696]
[WorldCat.org]
[DOI]
(I p)
Mark A Strauch, Benjamin G Bobay, John Cavanagh, Fude Yao, Angelo Wilson, Yoann Le Breton
Abh and AbrB control of Bacillus subtilis antimicrobial gene expression.
J Bacteriol: 2007, 189(21);7720-32
[PubMed:17720793]
[WorldCat.org]
[DOI]
(P p)
Akihito Ochiai, Takafumi Itoh, Akiko Kawamata, Wataru Hashimoto, Kousaku Murata
Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization.
Appl Environ Microbiol: 2007, 73(12);3803-13
[PubMed:17449691]
[WorldCat.org]
[DOI]
(P p)
Nicholas E E Allenby, Carys A Watts, Georg Homuth, Zoltán Prágai, Anil Wipat, Alan C Ward, Colin R Harwood
Phosphate starvation induces the sporulation killing factor of Bacillus subtilis.
J Bacteriol: 2006, 188(14);5299-303
[PubMed:16816204]
[WorldCat.org]
[DOI]
(P p)
Helga Westers, Peter G Braun, Lidia Westers, Haike Antelmann, Michael Hecker, Jan D H Jongbloed, Hirofumi Yoshikawa, Teruo Tanaka, Jan Maarten van Dijl, Wim J Quax
Genes involved in SkfA killing factor production protect a Bacillus subtilis lipase against proteolysis.
Appl Environ Microbiol: 2005, 71(4);1899-908
[PubMed:15812018]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, José Eduardo González-Pastor, Richard Losick
High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis.
J Bacteriol: 2005, 187(4);1357-68
[PubMed:15687200]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
José E González-Pastor, Errett C Hobbs, Richard Losick
Cannibalism by sporulating bacteria.
Science: 2003, 301(5632);510-3
[PubMed:12817086]
[WorldCat.org]
[DOI]
(I p)