SipW
- Description: signal peptidase I, required for biofilm formation
| Gene name | sipW |
| Synonyms | yqhE |
| Essential | no |
| Product | signal peptidase I |
| Function | biofilm formation |
| Regulation of this protein in SubtiPathways: Biofilm | |
| MW, pI | 20 kDa, 5.494 |
| Gene length, protein length | 570 bp, 190 aa |
| Immediate neighbours | tasA, yqxM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 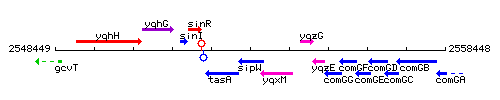
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU24630
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Cleavage of hydrophobic, N-terminal signal or leader sequences from secreted and periplasmic proteins (according to Swiss-Prot)
- Protein family: peptidase S26B family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: secreted (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P54506
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yunrong Chai, Thomas Norman, Roberto Kolter, Richard Losick
An epigenetic switch governing daughter cell separation in Bacillus subtilis.
Genes Dev: 2010, 24(8);754-65
[PubMed:20351052]
[WorldCat.org]
[DOI]
(I p)
Kazuo Kobayashi
SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 69(6);1399-410
[PubMed:18647168]
[WorldCat.org]
[DOI]
(I p)
Frances Chu, Daniel B Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter, Richard Losick
A novel regulatory protein governing biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 68(5);1117-27
[PubMed:18430133]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Frances Chu, Roberto Kolter, Richard Losick
Bistability and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 67(2);254-63
[PubMed:18047568]
[WorldCat.org]
[DOI]
(P p)
Mark A Strauch, Benjamin G Bobay, John Cavanagh, Fude Yao, Angelo Wilson, Yoann Le Breton
Abh and AbrB control of Bacillus subtilis antimicrobial gene expression.
J Bacteriol: 2007, 189(21);7720-32
[PubMed:17720793]
[WorldCat.org]
[DOI]
(P p)
Frances Chu, Daniel B Kearns, Steven S Branda, Roberto Kolter, Richard Losick
Targets of the master regulator of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2006, 59(4);1216-28
[PubMed:16430695]
[WorldCat.org]
[DOI]
(P p)
Steven S Branda, José Eduardo González-Pastor, Etienne Dervyn, S Dusko Ehrlich, Richard Losick, Roberto Kolter
Genes involved in formation of structured multicellular communities by Bacillus subtilis.
J Bacteriol: 2004, 186(12);3970-9
[PubMed:15175311]
[WorldCat.org]
[DOI]
(P p)
A G Stöver, A Driks
Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA.
J Bacteriol: 1999, 181(17);5476-81
[PubMed:10464223]
[WorldCat.org]
[DOI]
(P p)
M Serrano, R Zilhão, E Ricca, A J Ozin, C P Moran, A O Henriques
A Bacillus subtilis secreted protein with a role in endospore coat assembly and function.
J Bacteriol: 1999, 181(12);3632-43
[PubMed:10368135]
[WorldCat.org]
[DOI]
(P p)
H Tjalsma, A Bolhuis, M L van Roosmalen, T Wiegert, W Schumann, C P Broekhuizen, W J Quax, G Venema, S Bron, J M van Dijl
Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases.
Genes Dev: 1998, 12(15);2318-31
[PubMed:9694797]
[WorldCat.org]
[DOI]
(P p)