Difference between revisions of "SigW"
(→Extended information on the protein) |
|||
| Line 57: | Line 57: | ||
=== Additional information=== | === Additional information=== | ||
| − | + | * A mutation was found in this gene after evolution under relaxed selection for sporulation {{PubMed|21821766}} | |
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 141: | Line 139: | ||
<pubmed>9683469,16629676,17675383,11866510,9987136,10960106, </pubmed> | <pubmed>9683469,16629676,17675383,11866510,9987136,10960106, </pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed>11222589,14993308,16816000,17338440,12207695, 11454200,14563871,12076816, 19820159 </pubmed> | + | <pubmed>11222589,14993308,16816000,17338440,12207695, 11454200,14563871,12076816, 19820159 21821766</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:51, 15 August 2011
- Description: RNA polymerase ECF-type sigma factor SigW, activated by alkaline shock and by polymyxin B, vancomycin, cephalosporin C, D-cycloserine, and triton X-100
| Gene name | sigW |
| Synonyms | ybbL |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigW |
| Function | resistance against SdpC that functions in detoxification and/or production of antimicrobial compounds |
| Interactions involving this protein in SubtInteract: SigW | |
| MW, pI | 21 kDa, 8.614 |
| Gene length, protein length | 561 bp, 187 aa |
| Immediate neighbours | trnSL-Gln2, ybbM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 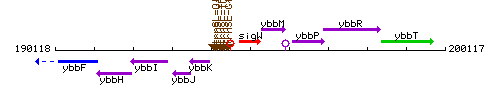
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, W, X, Y)
This gene is a member of the following regulons
The SigW regulon
The gene
Basic information
- Locus tag: BSU01730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: RsiW acts as antagonist of SigW (anti-SigW)
Database entries
- Structure:
- UniProt: Q45585
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Thomas Wiegert, University of Bayreuth, Germany Homepage
Your additional remarks
References
Additional publications: PubMed
The SigW regulon
Thorsten Mascher, Anna-Barbara Hachmann, John D Helmann
Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors.
J Bacteriol: 2007, 189(19);6919-27
[PubMed:17675383]
[WorldCat.org]
[DOI]
(P p)
Bronwyn G Butcher, John D Helmann
Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli.
Mol Microbiol: 2006, 60(3);765-82
[PubMed:16629676]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Phil A Kobel, Maud M Morshedi, Ming Fang Winston Wu, Chris Paddon, John D Helmann
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.
J Mol Biol: 2002, 316(3);443-57
[PubMed:11866510]
[WorldCat.org]
[DOI]
(P p)
M S Turner, J D Helmann
Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis.
J Bacteriol: 2000, 182(18);5202-10
[PubMed:10960106]
[WorldCat.org]
[DOI]
(P p)
X Huang, A Gaballa, M Cao, J D Helmann
Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W.
Mol Microbiol: 1999, 31(1);361-71
[PubMed:9987136]
[WorldCat.org]
[DOI]
(P p)
X Huang, K L Fredrick, J D Helmann
Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon.
J Bacteriol: 1998, 180(15);3765-70
[PubMed:9683469]
[WorldCat.org]
[DOI]
(P p)
Other original publications
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Jessica C Zweers, Thomas Wiegert, Jan Maarten van Dijl
Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis.
Appl Environ Microbiol: 2009, 75(23);7356-64
[PubMed:19820159]
[WorldCat.org]
[DOI]
(I p)
John D Helmann
Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds.
Sci Prog: 2006, 89(Pt 3-4);243-66
[PubMed:17338440]
[WorldCat.org]
[DOI]
(P p)
Craig D Ellermeier, Richard Losick
Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis.
Genes Dev: 2006, 20(14);1911-22
[PubMed:16816000]
[WorldCat.org]
[DOI]
(P p)
Mika Yoshimura, Kei Asai, Yoshito Sadaie, Hirofumi Yoshikawa
Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors.
Microbiology (Reading): 2004, 150(Pt 3);591-599
[PubMed:14993308]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Tamara Hoffmann, Ina Budde, Uwe Völker, Erhard Bremer
Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity.
J Bacteriol: 2003, 185(21);6358-70
[PubMed:14563871]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Tao Wang, Rick Ye, John D Helmann
Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons.
Mol Microbiol: 2002, 45(5);1267-76
[PubMed:12207695]
[WorldCat.org]
[DOI]
(P p)
Qiang Qian, Chien Y Lee, John D Helmann, Mark A Strauch
AbrB is a regulator of the sigma(W) regulon in Bacillus subtilis.
FEMS Microbiol Lett: 2002, 211(2);219-23
[PubMed:12076816]
[WorldCat.org]
[DOI]
(P p)
T Wiegert, G Homuth, S Versteeg, W Schumann
Alkaline shock induces the Bacillus subtilis sigma(W) regulon.
Mol Microbiol: 2001, 41(1);59-71
[PubMed:11454200]
[WorldCat.org]
[DOI]
(P p)
J Qiu, J D Helmann
The -10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function sigma factors sigma(X) and sigma(W).
J Bacteriol: 2001, 183(6);1921-7
[PubMed:11222589]
[WorldCat.org]
[DOI]
(P p)