Difference between revisions of "SigV"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 100: | Line 100: | ||
=== Additional information=== | === Additional information=== | ||
| + | * Expression of the [[SigV regulon]] in increased in ''[[ugtP]]'' mutants {{PubMed|22362028}} | ||
=Expression and regulation= | =Expression and regulation= | ||
| Line 135: | Line 136: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|20817771, 21926231,21856855}} | '''Additional publications:''' {{PubMed|20817771, 21926231,21856855}} | ||
| − | <pubmed>14993308,16274938,17675383,19745567</pubmed> | + | <pubmed>14993308,16274938,17675383,19745567 22362028</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:04, 4 March 2012
- Description: RNA polymerase ECF-type sigma factor SigV
| Gene name | sigV |
| Synonyms | |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigV |
| Function | response to lysozyme |
| Interactions involving this protein in SubtInteract: SigV | |
| MW, pI | 19 kDa, 9.037 |
| Gene length, protein length | 498 bp, 166 aa |
| Immediate neighbours | yrhO, rsiV |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 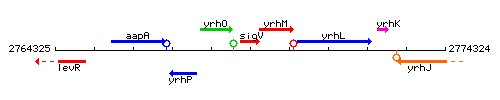
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The SigV regulon:
The gene
Basic information
- Locus tag: BSU27120
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O05404
- KEGG entry: [3]
- E.C. number:
Additional information
- Expression of the SigV regulon in increased in ugtP mutants PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Thomas Wiegert, University of Bayreuth, Germany Homepage
Your additional remarks
References
Additional publications: 21926231,21856855 PubMed
Satoshi Matsuoka, Minako Chiba, Yu Tanimura, Michihiro Hashimoto, Hiroshi Hara, Kouji Matsumoto
Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids.
Genes Genet Syst: 2011, 86(5);295-304
[PubMed:22362028]
[WorldCat.org]
[DOI]
(I p)
Michihiro Hashimoto, Hiroaki Takahashi, Yoshinori Hara, Hiroshi Hara, Kei Asai, Yoshito Sadaie, Kouji Matsumoto
Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with membranes of reduced phosphatidylglycerol content.
Genes Genet Syst: 2009, 84(3);191-8
[PubMed:19745567]
[WorldCat.org]
[DOI]
(P p)
Thorsten Mascher, Anna-Barbara Hachmann, John D Helmann
Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors.
J Bacteriol: 2007, 189(19);6919-27
[PubMed:17675383]
[WorldCat.org]
[DOI]
(P p)
Stephan Zellmeier, Claudia Hofmann, Sylvia Thomas, Thomas Wiegert, Wolfgang Schumann
Identification of sigma(V)-dependent genes of Bacillus subtilis.
FEMS Microbiol Lett: 2005, 253(2);221-9
[PubMed:16274938]
[WorldCat.org]
[DOI]
(P p)
Mika Yoshimura, Kei Asai, Yoshito Sadaie, Hirofumi Yoshikawa
Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors.
Microbiology (Reading): 2004, 150(Pt 3);591-599
[PubMed:14993308]
[WorldCat.org]
[DOI]
(P p)