Difference between revisions of "SigM"
| Line 13: | Line 13: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || responsible for intrinsic resistance against beta-lactam antibiotics | |style="background:#ABCDEF;" align="center"|'''Function''' || responsible for intrinsic resistance against beta-lactam antibiotics | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU09520 sigM] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SigM SigM] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SigM SigM] | ||
| Line 23: | Line 23: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yhdL]]'', ''[[yhdN]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yhdL]]'', ''[[yhdN]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU09520 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU09520 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU09520 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:sigM_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:sigM_context.gif]] | ||
Revision as of 12:26, 13 May 2013
- Description: RNA polymerase ECF-type sigma factor SigM
| Gene name | sigM |
| Synonyms | yhdM |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigM |
| Function | responsible for intrinsic resistance against beta-lactam antibiotics |
| Gene expression levels in SubtiExpress: sigM | |
| Interactions involving this protein in SubtInteract: SigM | |
| MW, pI | 19 kDa, 6.77 |
| Gene length, protein length | 489 bp, 163 aa |
| Immediate neighbours | yhdL, yhdN |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 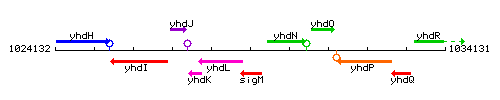
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against toxins/ antibiotics
This gene is a member of the following regulons
The SigM regulon
The gene
Basic information
- Locus tag: BSU09520
Phenotypes of a mutant
- SigM is essential for growth and survival in nutrient broth (NB) containing 1.4 M NaCl PubMed
- sigM mutants form aberrantly shaped cells, which swell and lyse spontaneously during growth in NB medium containing increased levels (0.35-0.7 M) of a wide range of different salts PubMed
- increased sensitivity towards beta-lactam antibiotics PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family, ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O07582
- KEGG entry: [3]
- E.C. number:
Additional information
- Expression of the SigM regulon in increased in ugtP mutants PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
The SigM regulon
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Thorsten Mascher, Anna-Barbara Hachmann, John D Helmann
Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors.
J Bacteriol: 2007, 189(19);6919-27
[PubMed:17675383]
[WorldCat.org]
[DOI]
(P p)
Adrian J Jervis, Penny D Thackray, Chris W Houston, Malcolm J Horsburgh, Anne Moir
SigM-responsive genes of Bacillus subtilis and their promoters.
J Bacteriol: 2007, 189(12);4534-8
[PubMed:17434969]
[WorldCat.org]
[DOI]
(P p)
Other original publications