Difference between revisions of "SigA"
(→References) |
(→Extended information on the protein) |
||
| Line 100: | Line 100: | ||
** [[BsrB]]-[[SigA]]-[[RNA polymerase]] | ** [[BsrB]]-[[SigA]]-[[RNA polymerase]] | ||
| − | * '''[[Localization]]:''' | + | * '''[[Localization]]:''' |
| + | ** cytoplasm {{PubMed|25313396}} | ||
=== Database entries === | === Database entries === | ||
Revision as of 16:40, 27 October 2014
- Description: RNA polymerase major sigma factor SigA
| Gene name | sigA |
| Synonyms | rpoD, crsA |
| Essential | yes PubMed |
| Product | RNA polymerase major sigma factor SigA |
| Function | transcription |
| Gene expression levels in SubtiExpress: sigA | |
| Interactions involving this protein in SubtInteract: SigA | |
| MW, pI | 42 kDa, 4.634 |
| Gene length, protein length | 1113 bp, 371 aa |
| Immediate neighbours | cccA, dnaG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 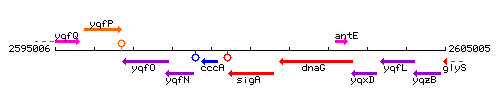
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed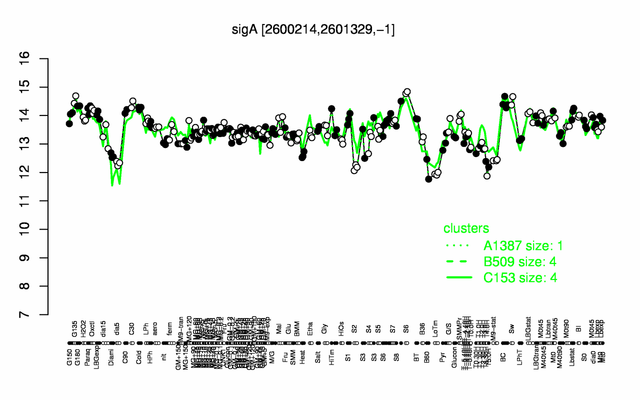
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25200
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU25200
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm PubMed
Database entries
- BsubCyc: BSU25200
- Structure:
- UniProt: P06224
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- An antisense RNA is predicted for sigA PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 802 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1811 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 1136 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2062 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1407 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
Benedikt Steuten, Sabine Schneider, Rolf Wagner
6S RNA: recent answers--future questions.
Mol Microbiol: 2014, 91(4);641-8
[PubMed:24308327]
[WorldCat.org]
[DOI]
(I p)
Lakshminarayan M Iyer, L Aravind
Insights from the architecture of the bacterial transcription apparatus.
J Struct Biol: 2012, 179(3);299-319
[PubMed:22210308]
[WorldCat.org]
[DOI]
(I p)
Original publications
Yong Heon Lee, John D Helmann
Mutations in the primary sigma factor σA and termination factor rho that reduce susceptibility to cell wall antibiotics.
J Bacteriol: 2014, 196(21);3700-11
[PubMed:25112476]
[WorldCat.org]
[DOI]
(I p)
Hsin-Yi Yeh, Hsiu-Ting Hsu, Tsung-Ching Chen, Kuei-Min Chung, Kung-Ming Liou, Ban-Yang Chang
The reduction in σ-promoter recognition flexibility as induced by core RNAP is required for σ to discern the optimal promoter spacing.
Biochem J: 2013, 455(2);185-93
[PubMed:23875654]
[WorldCat.org]
[DOI]
(I p)
Benedikt M Beckmann, Philipp G Hoch, Manja Marz, Dagmar K Willkomm, Margarita Salas, Roland K Hartmann
A pRNA-induced structural rearrangement triggers 6S-1 RNA release from RNA polymerase in Bacillus subtilis.
EMBO J: 2012, 31(7);1727-38
[PubMed:22333917]
[WorldCat.org]
[DOI]
(I p)
Hsin-Yi Yeh, Tsung-Ching Chen, Kung-Ming Liou, Hsiu-Ting Hsu, Kuei-Min Chung, Li-Ling Hsu, Ban-Yang Chang
The core-independent promoter-specific interaction of primary sigma factor.
Nucleic Acids Res: 2011, 39(3);913-25
[PubMed:20935043]
[WorldCat.org]
[DOI]
(I p)
Shu Ishikawa, Taku Oshima, Ken Kurokawa, Yoko Kusuya, Naotake Ogasawara
RNA polymerase trafficking in Bacillus subtilis cells.
J Bacteriol: 2010, 192(21);5778-87
[PubMed:20817769]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Elecia B Johnston, Peter J Lewis, Renate Griffith
The interaction of Bacillus subtilis sigmaA with RNA polymerase.
Protein Sci: 2009, 18(11);2287-97
[PubMed:19735077]
[WorldCat.org]
[DOI]
(I p)
Xiao Yang, Seeseei Molimau, Geoff P Doherty, Elecia B Johnston, Jon Marles-Wright, Rosalba Rothnagel, Ben Hankamer, Richard J Lewis, Peter J Lewis
The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA.
EMBO Rep: 2009, 10(9);997-1002
[PubMed:19680289]
[WorldCat.org]
[DOI]
(I p)
Steve D Seredick, George B Spiegelman
The Bacillus subtilis response regulator Spo0A stimulates sigmaA-dependent transcription prior to the major energetic barrier.
J Biol Chem: 2004, 279(17);17397-403
[PubMed:14976210]
[WorldCat.org]
[DOI]
(P p)
Amrita Kumar, James A Brannigan, Charles P Moran
Alpha-helix E of Spo0A is required for sigmaA- but not for sigmaH-dependent promoter activation in Bacillus subtilis.
J Bacteriol: 2004, 186(4);1078-83
[PubMed:14762002]
[WorldCat.org]
[DOI]
(P p)
Amrita Kumar, Cindy Buckner Starke, Mark DeZalia, Charles P Moran
Surfaces of Spo0A and RNA polymerase sigma factor A that interact at the spoIIG promoter in Bacillus subtilis.
J Bacteriol: 2004, 186(1);200-6
[PubMed:14679239]
[WorldCat.org]
[DOI]
(P p)
Claudia Rollenhagen, Haike Antelmann, Janine Kirstein, Olivier Delumeau, Michael Hecker, Michael D Yudkin
Binding of sigma(A) and sigma(B) to core RNA polymerase after environmental stress in Bacillus subtilis.
J Bacteriol: 2003, 185(1);35-40
[PubMed:12486038]
[WorldCat.org]
[DOI]
(P p)
Hanne Jarmer, Thomas S Larsen, Anders Krogh, Hans Henrik Saxild, Søren Brunak, Steen Knudsen
Sigma A recognition sites in the Bacillus subtilis genome.
Microbiology (Reading): 2001, 147(Pt 9);2417-2424
[PubMed:11535782]
[WorldCat.org]
[DOI]
(P p)
J Qiu, J D Helmann
Adenines at -11, -9 and -8 play a key role in the binding of Bacillus subtilis Esigma(A) RNA polymerase to -10 region single-stranded DNA.
Nucleic Acids Res: 1999, 27(23);4541-6
[PubMed:10556308]
[WorldCat.org]
[DOI]
(P p)
J Ju, T Mitchell, H Peters, W G Haldenwang
Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation.
J Bacteriol: 1999, 181(16);4969-77
[PubMed:10438769]
[WorldCat.org]
[DOI]
(P p)
C M Buckner, G Schyns, C P Moran
A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation.
J Bacteriol: 1998, 180(14);3578-83
[PubMed:9658000]
[WorldCat.org]
[DOI]
(P p)
G Schyns, C M Buckner, C P Moran
Activation of the Bacillus subtilis spoIIG promoter requires interaction of Spo0A and the sigma subunit of RNA polymerase.
J Bacteriol: 1997, 179(17);5605-8
[PubMed:9287022]
[WorldCat.org]
[DOI]
(P p)
X Huang, F J Lopez de Saro, J D Helmann
sigma factor mutations affecting the sequence-selective interaction of RNA polymerase with -10 region single-stranded DNA.
Nucleic Acids Res: 1997, 25(13);2603-9
[PubMed:9185571]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, J D Helmann
RNA polymerase sigma factor determines start-site selection but is not required for upstream promoter element activation on heteroduplex (bubble) templates.
Proc Natl Acad Sci U S A: 1997, 94(10);4982-7
[PubMed:9144176]
[WorldCat.org]
[DOI]
(P p)
J D Helmann
Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA.
Nucleic Acids Res: 1995, 23(13);2351-60
[PubMed:7630711]
[WorldCat.org]
[DOI]
(P p)
Y L Juang, J D Helmann
Pathway of promoter melting by Bacillus subtilis RNA polymerase at a stable RNA promoter: effects of temperature, delta protein, and sigma factor mutations.
Biochemistry: 1995, 34(26);8465-73
[PubMed:7599136]
[WorldCat.org]
[DOI]
(P p)
S E Aiyar, Y L Juang, J D Helmann, P L deHaseth
Mutations in sigma factor that affect the temperature dependence of transcription from a promoter, but not from a mismatch bubble in double-stranded DNA.
Biochemistry: 1994, 33(38);11501-6
[PubMed:7918363]
[WorldCat.org]
[DOI]
(P p)
J C Rong, J D Helmann
Genetic and physiological studies of Bacillus subtilis sigma A mutants defective in promoter melting.
J Bacteriol: 1994, 176(17);5218-24
[PubMed:8071196]
[WorldCat.org]
[DOI]
(P p)
Y L Juang, J D Helmann
A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids.
J Mol Biol: 1994, 235(5);1470-88
[PubMed:8107087]
[WorldCat.org]
[DOI]
(P p)
H L Carter, L F Wang, R H Doi, C P Moran
rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis.
J Bacteriol: 1988, 170(4);1617-21
[PubMed:3127379]
[WorldCat.org]
[DOI]
(P p)
C W Price, R H Doi
Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation.
Mol Gen Genet: 1985, 201(1);88-95
[PubMed:2997585]
[WorldCat.org]
[DOI]
(P p)