Difference between revisions of "ScoC"
| Line 41: | Line 41: | ||
{{SubtiWiki regulon|[[SalA regulon]]}}, | {{SubtiWiki regulon|[[SalA regulon]]}}, | ||
{{SubtiWiki regulon|[[SenS regulon]]}} | {{SubtiWiki regulon|[[SenS regulon]]}} | ||
| + | |||
| + | =The [[ScoC regulon]]= | ||
=The gene= | =The gene= | ||
Revision as of 20:36, 20 December 2010
- Description: transcriptional repressor of genes expressed in the transition phase
| Gene name | scoC |
| Synonyms | hpr, catA |
| Essential | no |
| Product | transcriptional repressor (MarR family) |
| Function | transition state regulator |
| Regulation of this protein in SubtiPathways: Biofilm, Alternative nitrogen sources | |
| MW, pI | 23 kDa, 5.188 |
| Gene length, protein length | 609 bp, 203 aa |
| Immediate neighbours | yhaI, yhaH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 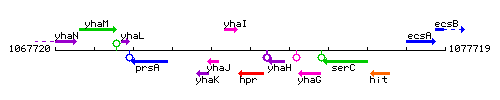
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control, transition state regulators
This gene is a member of the following regulons
AbrB regulon, SalA regulon, SenS regulon
The ScoC regulon
The gene
Basic information
- Locus tag: BSU09990
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Genes controlled by ScoC
repressed: aprE, bacA-bacB-bacC-bacD-bacE-bacF PubMed, hag PubMed, flgM PubMed
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 2FXA
- UniProt: P11065
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: scoC PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Bindiya Kaushal, Salbi Paul, F Marion Hulett
Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC.
J Bacteriol: 2010, 192(12);3103-13
[PubMed:20382764]
[WorldCat.org]
[DOI]
(I p)
Prashant Kodgire, K Krishnamurthy Rao
A dual mode of regulation of flgM by ScoC in Bacillus subtilis.
Can J Microbiol: 2009, 55(8);983-9
[PubMed:19898538]
[WorldCat.org]
[DOI]
(I p)
Takashi Inaoka, Guojun Wang, Kozo Ochi
ScoC regulates bacilysin production at the transcription level in Bacillus subtilis.
J Bacteriol: 2009, 191(23);7367-71
[PubMed:19801406]
[WorldCat.org]
[DOI]
(I p)
Prashant Kodgire, K Krishnamurthy Rao
hag expression in Bacillus subtilis is both negatively and positively regulated by ScoC.
Microbiology (Reading): 2009, 155(Pt 1);142-149
[PubMed:19118355]
[WorldCat.org]
[DOI]
(P p)
Prashant Kodgire, Madhulika Dixit, K Krishnamurthy Rao
ScoC and SinR negatively regulate epr by corepression in Bacillus subtilis.
J Bacteriol: 2006, 188(17);6425-8
[PubMed:16923912]
[WorldCat.org]
[DOI]
(P p)
Eiji Kawachi, Sadanobu Abe, Teruo Tanaka
Inhibition of Bacillus subtilis scoC expression by multicopy senS.
J Bacteriol: 2005, 187(24);8526-30
[PubMed:16321961]
[WorldCat.org]
[DOI]
(P p)
Mitsuo Ogura, Atsushi Matsuzawa, Hirofumi Yoshikawa, Teruo Tanaka
Bacillus subtilis SalA (YbaL) negatively regulates expression of scoC, which encodes the repressor for the alkaline exoprotease gene, aprE.
J Bacteriol: 2004, 186(10);3056-64
[PubMed:15126467]
[WorldCat.org]
[DOI]
(P p)
Alejandro Sánchez, Jorge Olmos
Bacillus subtilis transcriptional regulators interaction.
Biotechnol Lett: 2004, 26(5);403-7
[PubMed:15104138]
[WorldCat.org]
[DOI]
(P p)
Sasha H Shafikhani, Esperanza Núñez, Terrance Leighton
ScoC mediates catabolite repression of sporulation in Bacillus subtilis.
Curr Microbiol: 2003, 47(4);327-36
[PubMed:14629015]
[WorldCat.org]
[DOI]
(P p)
A Koide, M Perego, J A Hoch
ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis.
J Bacteriol: 1999, 181(13);4114-7
[PubMed:10383984]
[WorldCat.org]
[DOI]
(P p)
P T Kallio, J E Fagelson, J A Hoch, M A Strauch
The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein.
J Biol Chem: 1991, 266(20);13411-7
[PubMed:1906467]
[WorldCat.org]
(P p)
B C Dowds, J A Hoch
Regulation of the oxidative stress response by the hpr gene in Bacillus subtilis.
J Gen Microbiol: 1991, 137(5);1121-5
[PubMed:1907636]
[WorldCat.org]
[DOI]
(P p)
M A Strauch, G B Spiegelman, M Perego, W C Johnson, D Burbulys, J A Hoch
The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein.
EMBO J: 1989, 8(5);1615-21
[PubMed:2504584]
[WorldCat.org]
[DOI]
(P p)
M Perego, J A Hoch
Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis.
J Bacteriol: 1988, 170(6);2560-7
[PubMed:3131303]
[WorldCat.org]
[DOI]
(P p)