Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' two-component | + | * '''Description:''' two-component response regulator, regulation of degradative enzyme and genetic competence, involved in activation of capsule biosynthetic operon expression (together with [[SwrAA]]), non-phosphorylated DegU is required for swarming motility [http://www3.interscience.wiley.com/journal/122328658/abstract PubMed] <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''degU'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''sacU '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''sacU, iep '' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || two-component | + | |style="background:#ABCDEF;" align="center"| '''Product''' || two-component response regulator |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of degradative enzyme and genetic competence | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of degradative enzyme and genetic competence | ||
| + | of genetic competence | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 25 kDa, 5.602 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 687 bp, 229 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yviA]]'', ''[[degS]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15566]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:degU_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 36: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU35490 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 43: | Line 44: | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/degSU.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/degSU.html] | ||
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10393] |
=== Additional information=== | === Additional information=== | ||
| Line 52: | Line 53: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' transcription activator of degradative enzyme genes (''[[sacB]]'' and ''[[degQ]]'') and of ''[[comK]]'' expression |
* '''Protein family:''' | * '''Protein family:''' | ||
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| + | |||
| + | === Genes controlled by DegU === | ||
| + | |||
| + | * '''Activation by DegU-P:''' ''[[degS]]-[[degU]]'' | ||
| + | |||
| + | * '''Activation by DegU:''' ''[[epr]]'' [http://www3.interscience.wiley.com/journal/122328658/abstract PubMed] | ||
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 64: | Line 71: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | + | * '''Modification:''' phosphorylated by [[DegS]] on Asp-56 [http://www.ncbi.nlm.nih.gov/sites/entrez/1901568 PubMed], this modulates DNA-binding activity | |
| − | |||
| − | |||
| − | |||
| − | * '''Modification:''' | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' |
| − | * '''Interactions:''' [[ | + | * '''Interactions:''' [[DegU]]-[[RapG]], [[DegS]]-[[DegU]] |
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasm (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 82: | Line 85: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P13800 P13800] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU35490] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 97: | Line 100: | ||
* '''Regulation:''' induced in the presence of ammonium ([[TnrA]]) | * '''Regulation:''' induced in the presence of ammonium ([[TnrA]]) | ||
| + | ** repressed in the absence of good nitrogen sources (glutamine or ammonium) ([[TnrA]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/18502860 PubMed] | ||
* '''Regulatory mechanism:''' [[TnrA]]: repression, [[DegU]]: positive autoregulation | * '''Regulatory mechanism:''' [[TnrA]]: repression, [[DegU]]: positive autoregulation | ||
| + | ** [[TnrA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/18502860 PubMed] | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 117: | Line 122: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | |||
| − | |||
=Your additional remarks= | =Your additional remarks= | ||
| Line 124: | Line 127: | ||
=References= | =References= | ||
| − | <pubmed>17850253,12471443,14563871,1459944, | + | <pubmed>17850253,12471443,8878039,14563871,1901568,1321152,1459944,18194340,10908654,18502860,10094672,19389763,18414485,19420703,2428811,7746142,,18502860, 19389763, 19416356, 19118340 </pubmed> |
| − | |||
| − | |||
Revision as of 09:03, 13 June 2009
- Description: two-component response regulator, regulation of degradative enzyme and genetic competence, involved in activation of capsule biosynthetic operon expression (together with SwrAA), non-phosphorylated DegU is required for swarming motility PubMed
| Gene name | degU |
| Synonyms | sacU, iep |
| Essential | no |
| Product | two-component response regulator |
| Function | regulation of degradative enzyme and genetic competence
of genetic competence |
| MW, pI | 25 kDa, 5.602 |
| Gene length, protein length | 687 bp, 229 aa |
| Immediate neighbours | yviA, degS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 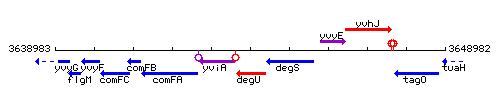
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU35490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activator of degradative enzyme genes (sacB and degQ) and of comK expression
- Protein family:
- Paralogous protein(s):
Genes controlled by DegU
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P13800
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: induced in the presence of ammonium (TnrA)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Keitarou Kimura, Lam-Son Phan Tran, Thi-Huyen Do, Yoshifumi Itoh
Expression of the pgsB encoding the poly-gamma-DL-glutamate synthetase of Bacillus subtilis (natto).
Biosci Biotechnol Biochem: 2009, 73(5);1149-55
[PubMed:19420703]
[WorldCat.org]
[DOI]
(I p)
Monica Gupta, Kestur Krishnamurthy Rao
Epr plays a key role in DegU-mediated swarming motility of Bacillus subtilis.
FEMS Microbiol Lett: 2009, 295(2);187-94
[PubMed:19416356]
[WorldCat.org]
[DOI]
(I p)
Cecilia Osera, Giuseppe Amati, Cinzia Calvio, Alessandro Galizzi
SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 7);2282-2287
[PubMed:19389763]
[WorldCat.org]
[DOI]
(P p)
Ewan J Murray, Taryn B Kiley, Nicola R Stanley-Wall
A pivotal role for the response regulator DegU in controlling multicellular behaviour.
Microbiology (Reading): 2009, 155(Pt 1);1-8
[PubMed:19118340]
[WorldCat.org]
[DOI]
(P p)
Ayako Yasumura, Sadanobu Abe, Teruo Tanaka
Involvement of nitrogen regulation in Bacillus subtilis degU expression.
J Bacteriol: 2008, 190(15);5162-71
[PubMed:18502860]
[WorldCat.org]
[DOI]
(I p)
Jan-Willem Veening, Oleg A Igoshin, Robyn T Eijlander, Reindert Nijland, Leendert W Hamoen, Oscar P Kuipers
Transient heterogeneity in extracellular protease production by Bacillus subtilis.
Mol Syst Biol: 2008, 4;184
[PubMed:18414485]
[WorldCat.org]
[DOI]
(I p)
Kensuke Tsukahara, Mitsuo Ogura
Characterization of DegU-dependent expression of bpr in Bacillus subtilis.
FEMS Microbiol Lett: 2008, 280(1);8-13
[PubMed:18194340]
[WorldCat.org]
[DOI]
(P p)
Kazuo Kobayashi
Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2007, 66(2);395-409
[PubMed:17850253]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Tamara Hoffmann, Ina Budde, Uwe Völker, Erhard Bremer
Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity.
J Bacteriol: 2003, 185(21);6358-70
[PubMed:14563871]
[WorldCat.org]
[DOI]
(P p)
U Mäder, H Antelmann, T Buder, M K Dahl, M Hecker, G Homuth
Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics.
Mol Genet Genomics: 2002, 268(4);455-67
[PubMed:12471443]
[WorldCat.org]
[DOI]
(P p)
L W Hamoen, A F Van Werkhoven, G Venema, D Dubnau
The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2000, 97(16);9246-51
[PubMed:10908654]
[WorldCat.org]
[DOI]
(P p)
C Fabret, V A Feher, J A Hoch
Two-component signal transduction in Bacillus subtilis: how one organism sees its world.
J Bacteriol: 1999, 181(7);1975-83
[PubMed:10094672]
[WorldCat.org]
[DOI]
(P p)
J Hahn, A Luttinger, D Dubnau
Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis.
Mol Microbiol: 1996, 21(4);763-75
[PubMed:8878039]
[WorldCat.org]
[DOI]
(P p)
B J Haijema, L W Hamoen, J Kooistra, G Venema, D van Sinderen
Expression of the ATP-dependent deoxyribonuclease of Bacillus subtilis is under competence-mediated control.
Mol Microbiol: 1995, 15(2);203-11
[PubMed:7746142]
[WorldCat.org]
[DOI]
(P p)
K Mukai, M Kawata-Mukai, T Tanaka
Stabilization of phosphorylated Bacillus subtilis DegU by DegR.
J Bacteriol: 1992, 174(24);7954-62
[PubMed:1459944]
[WorldCat.org]
[DOI]
(P p)
M K Dahl, T Msadek, F Kunst, G Rapoport
The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis.
J Biol Chem: 1992, 267(20);14509-14
[PubMed:1321152]
[WorldCat.org]
(P p)
M K Dahl, T Msadek, F Kunst, G Rapoport
Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase.
J Bacteriol: 1991, 173(8);2539-47
[PubMed:1901568]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes.
J Bacteriol: 1986, 168(1);380-8
[PubMed:2428811]
[WorldCat.org]
[DOI]
(P p)