Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' MreD is a cell shape determining protein and is associated with the MreB cytoskeleton in B. subtilis and other rod shaped bacteria. <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''mreD'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''rodB '' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || cell-shape determining protein |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || cell-shape determation |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 19 kDa, 7.906 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 516 bp, 172 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[minC]]'', ''[[mreC]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB14761]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:mreD_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU28010 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | + | the phenotype of ''mreD'' is similar to that of ''[[mreC]]''. ''mreD'' is essential under normal conditions [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed]. Depletion of MreD leads to a progressive increase in the width and a decrease in the length of the cell and cells become lytic. In the depletion strain, lysis can be prevented and cell growth, but not cell shape, can be recovered by inculaion of Magnesium in the media. This shape defect is consistent with a role for ''mreD'' in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation. |
| + | |||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/mreBCD-minCD.html] |
| + | |||
| + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10328] | ||
| + | |||
| + | = Additional information= | ||
| + | |||
| + | |||
| + | ===Function=== | ||
| − | + | MreD functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. As a transmembrane protein MreD is thought to provide a patch on the membrane that MreB inteacts with. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell. | |
| − | === | + | ===''mreD'' and the mapping of ''mre-min'' operon=== |
| + | The ''mre-min'' operon was first sequenced by levin et al in 1992. The temperature sentitive ''rodB1'' mutation mapperd to that region. | ||
=The protein= | =The protein= | ||
| Line 54: | Line 63: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' None/ structural |
| − | * '''Protein family:''' | + | * '''Protein family:''' mreD family (according to Swiss-Prot) COG2891 |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' None |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 64: | Line 73: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''Domains:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' |
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' |
| − | * '''Interactions:''' | + | * '''Interactions:''' |
| − | * '''Localization:''' | + | * '''Localization:''' |
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' None |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/Q01467 Q01467] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU28010] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 97: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[mreB]]''-''[[mreC]]''-'''mreD'''-''[[minC]]''-''[[minD]]'' |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigH]] [http://www.ncbi.nlm.nih.gov/sites/entrez/8459776 PubMed] |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 99: | Line 107: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant: '' | + | * '''Mutant:''' A conditional mutant with an inframe deletion of ''mreD'' complemented by a xylose inducible copy at an ectopic locus (strain named 4352) is avaliable from the Errington Group. |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 109: | Line 117: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| − | * '''GFP fusion:''' | + | * '''GFP fusion:''' A functional N-terminal GFP fusion has been made where the fusion protein is the only copy of the gene in the cell: strain 3416 [http://www.ncbi.nlm.nih.gov/pubmed/16101995 PubMed]. |
| + | |||
* '''two-hybrid system:''' | * '''two-hybrid system:''' | ||
| Line 116: | Line 125: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Peter Graumann]], Freiburg University, Germany [http://www.biologie.uni-freiburg.de/data/bio2/graumann/index.htm homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 121: | Line 132: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>1400224,18156271,16101995,, </pubmed> |
| − | |||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 09:00, 13 June 2009
- Description: MreD is a cell shape determining protein and is associated with the MreB cytoskeleton in B. subtilis and other rod shaped bacteria.
| Gene name | mreD |
| Synonyms | rodB |
| Essential | yes |
| Product | cell-shape determining protein |
| Function | cell-shape determation |
| MW, pI | 19 kDa, 7.906 |
| Gene length, protein length | 516 bp, 172 aa |
| Immediate neighbours | minC, mreC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 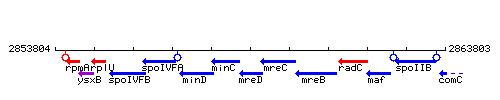
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28010
Phenotypes of a mutant
the phenotype of mreD is similar to that of mreC. mreD is essential under normal conditions PubMed. Depletion of MreD leads to a progressive increase in the width and a decrease in the length of the cell and cells become lytic. In the depletion strain, lysis can be prevented and cell growth, but not cell shape, can be recovered by inculaion of Magnesium in the media. This shape defect is consistent with a role for mreD in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Function
MreD functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. As a transmembrane protein MreD is thought to provide a patch on the membrane that MreB inteacts with. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell.
mreD and the mapping of mre-min operon
The mre-min operon was first sequenced by levin et al in 1992. The temperature sentitive rodB1 mutation mapperd to that region.
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: None/ structural
- Protein family: mreD family (according to Swiss-Prot) COG2891
- Paralogous protein(s): None
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: None
- Swiss prot entry: Q01467
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: A conditional mutant with an inframe deletion of mreD complemented by a xylose inducible copy at an ectopic locus (strain named 4352) is avaliable from the Errington Group.
- Expression vector:
- lacZ fusion:
- GFP fusion: A functional N-terminal GFP fusion has been made where the fusion protein is the only copy of the gene in the cell: strain 3416 PubMed.
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Alex Formstone, Rut Carballido-López, Philippe Noirot, Jeffery Errington, Dirk-Jan Scheffers
Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis.
J Bacteriol: 2008, 190(5);1812-21
[PubMed:18156271]
[WorldCat.org]
[DOI]
(I p)
Mark Leaver, Jeff Errington
Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis.
Mol Microbiol: 2005, 57(5);1196-209
[PubMed:16101995]
[WorldCat.org]
[DOI]
(P p)
P A Levin, P S Margolis, P Setlow, R Losick, D Sun
Identification of Bacillus subtilis genes for septum placement and shape determination.
J Bacteriol: 1992, 174(21);6717-28
[PubMed:1400224]
[WorldCat.org]
[DOI]
(P p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed