Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' cell-division protein | + | * '''Description:''' cell-division initiation protein (septum formation) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''ftsZ'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ts-1 '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || yes [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || cell-division protein | + | |style="background:#ABCDEF;" align="center"| '''Product''' || cell-division initiation protein (septum formation) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || formation of Z-ring | |style="background:#ABCDEF;" align="center"|'''Function''' || formation of Z-ring | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 40 kDa, 4.814 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1146 bp, 382 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ftsA]]'', ''[[bpr]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13402]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
|- | |- | ||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:ftsZ_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 37: | Line 37: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU15290 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 47: | Line 47: | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsAZ.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsAZ.html] | ||
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10232] |
=== Additional information=== | === Additional information=== | ||
| Line 58: | Line 58: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' ftsZ family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 74: | Line 74: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[FtsZ]]-[[FtsA]] | + | * '''Interactions:''' [[FtsZ]]-[[EzrA]], [[FtsZ]]-[[SpoIIE]], [[FtsZ]]-[[FtsA]], [[FtsZ]]-[[FtsZ]], [[MciZ]]-[[FtsZ]], [[FtsZ]]-[[SepF]] |
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasm (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2VAM 2VAM], [http://www.rcsb.org/pdb/explore.do?structureId=2RHL 2RHL] (dimer with GDP) |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P17865 P17865] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU15290] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 92: | Line 92: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' |
| − | * '''Regulation:''' | + | * '''Regulation:''' |
** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/7592498 PubMed] | ** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/7592498 PubMed] | ||
| Line 102: | Line 102: | ||
** [[AbrB]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/7592498 PubMed] | ** [[AbrB]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/7592498 PubMed] | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 114: | Line 114: | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | * '''two-hybrid system:''' | + | * '''two-hybrid system:''' |
| − | * '''Antibody:''' | + | * '''Antibody:''' available in the [[Jeff Errington]] lab |
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Imrich Barak]], Slovak Academy of Science, Bratislava, Slovakia [http://imb.savba.sk/~barak/ homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 124: | Line 126: | ||
=References= | =References= | ||
| − | <pubmed>7592498, </pubmed> | + | <pubmed>7592498, 19136590 , 19429628, 19141479 </pubmed> |
| − | |||
Revision as of 10:51, 12 June 2009
- Description: cell-division initiation protein (septum formation)
| Gene name | ftsZ |
| Synonyms | ts-1 |
| Essential | yes PubMed |
| Product | cell-division initiation protein (septum formation) |
| Function | formation of Z-ring |
| MW, pI | 40 kDa, 4.814 |
| Gene length, protein length | 1146 bp, 382 aa |
| Immediate neighbours | ftsA, bpr |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 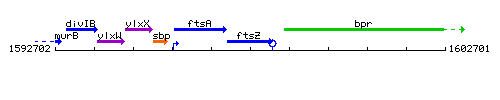
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15290
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsZ family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Swiss prot entry: P17865
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Jeff Errington lab
Labs working on this gene/protein
Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
Your additional remarks
References
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
James A Gregory, Eric C Becker, Kit Pogliano
Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division.
Genes Dev: 2008, 22(24);3475-88
[PubMed:19141479]
[WorldCat.org]
[DOI]
(P p)
Daniel P Haeusser, Amy H Lee, Richard B Weart, Petra Anne Levin
ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity.
J Bacteriol: 2009, 191(6);1986-91
[PubMed:19136590]
[WorldCat.org]
[DOI]
(I p)
M A Strauch
Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote.
J Bacteriol: 1995, 177(23);6999-7002
[PubMed:7592498]
[WorldCat.org]
[DOI]
(P p)