Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' ATP-dependent Clp protease, ATPase subunit <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''clpC'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''mecB '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || ATP-dependent Clp protease, ATPase subunit |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation |
| + | positive regulator of autolysin ([[LytC]] and [[LytD]]) synthesis | ||
|- | |- | ||
| − | | | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 89 kDa, 5.746 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| ''' | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2430 bp, 810 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| ''' | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[mcsB]]'', ''[[radA]]'' |
|- | |- | ||
| − | |style="background:# | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB14213]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | + | |colspan="2" | '''Genetic context''' <br/> [[Image:clpC_context.gif]] | |
| − | |||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 31: | Line 30: | ||
__TOC__ | __TOC__ | ||
| − | <br/><br/> | + | <br/><br/><br/><br/> |
=The gene= | =The gene= | ||
| Line 37: | Line 36: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU00860 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 43: | Line 42: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ctsR-mcsAB-clpC-radA-yacK.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10148] |
=== Additional information=== | === Additional information=== | ||
| Line 54: | Line 53: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' ATPase/chaperone |
| − | * '''Protein family:''' | + | * '''Protein family:''' mecA family (according to Swiss-Prot) clpA/clpB family. ClpC subfamily (according to Swiss-Prot), AAA+ -type ATPase (IPR013093) [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR013093 InterPro] (PF07724) [http://pfam.sanger.ac.uk/family?acc=PF07724 PFAM] |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[ClpE]], [[ClpX]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 64: | Line 63: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''Domains:''' AAA-ATPase [http://pfam.sanger.ac.uk/family?acc=PF07724 PFAM] |
* '''Modification:''' | * '''Modification:''' | ||
| Line 72: | Line 71: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[YpbH]]-[[ClpC]], [[McsB]]-[[ClpC]], [[ClpC]]-[[MecA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/10447896 PubMed], [[ClpC]]-[[ClpP]] |
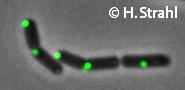
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heatshock, colocalization with [[ClpP]] [http://www.ncbi.nlm.nih.gov/pubmed/18786145 Pubmed] |
| + | [[File:ClpC.jpg ]] | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2K77 2K77] (N-terminal domain) |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P37571 P37571] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU00860] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 88: | ||
=== Additional information=== | === Additional information=== | ||
| + | :* subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[ctsR]]-[[mcsA]]-[[mcsB]]-[[clpC]]-[[radA]]-[[disA]]'' [http://www.ncbi.nlm.nih.gov/pubmed/8793870 PubMed] |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/pubmed/8793870 PubMed], [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/8793870 PubMed1] [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed2] |
| − | * '''Regulation:''' | + | * '''Regulation:''' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed], induced by heat ([[CtsR]]) [http://www.ncbi.nlm.nih.gov/pubmed/9987115 PubMed] |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' [[CtsR]]: transcription repression [http://www.ncbi.nlm.nih.gov/pubmed/9987115 PubMed1], [http://www.ncbi.nlm.nih.gov/pubmed/11179229 PubMed2], [http://www.ncbi.nlm.nih.gov/pubmed/16163393 PubMed3], [http://www.ncbi.nlm.nih.gov/pubmed/17380125 PubMed4] |
| − | * '''Additional information:''' | + | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] |
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' ''clpC::tet'' available from the [http://subtiwiki.uni-goettingen.de/wiki/index.php/Leendert_Hamoen Hamoen]] Lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 108: | Line 109: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| − | * '''GFP fusion:''' | + | * '''GFP fusion:''' C-terminal GFP fusions (single copy, also as CFP and YFP variants) available from the [http://subtiwiki.uni-goettingen.de/wiki/index.php/Leendert_Hamoen Hamoen]] Lab |
* '''two-hybrid system:''' | * '''two-hybrid system:''' | ||
| Line 115: | Line 116: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | [[Leendert Hamoen]], Newcastle University, UK [http://www.ncl.ac.uk/camb/staff/profile/l.hamoen homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
=References= | =References= | ||
| + | |||
| + | <pubmed> 11544224, 17981983, 14763982, 18786145, 19361434 </pubmed> | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 11:41, 11 June 2009
- Description: ATP-dependent Clp protease, ATPase subunit
| Gene name | clpC |
| Synonyms | mecB |
| Essential | no |
| Product | ATP-dependent Clp protease, ATPase subunit |
| Function | protein degradation |
| MW, pI | 89 kDa, 5.746 |
| Gene length, protein length | 2430 bp, 810 aa |
| Immediate neighbours | mcsB, radA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 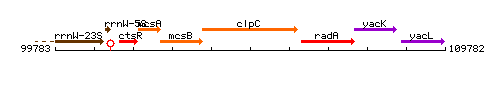
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU00860
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
- Protein family: mecA family (according to Swiss-Prot) clpA/clpB family. ClpC subfamily (according to Swiss-Prot), AAA+ -type ATPase (IPR013093) InterPro (PF07724) PFAM
Extended information on the protein
- Kinetic information:
- Domains: AAA-ATPase PFAM
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heatshock, colocalization with ClpP Pubmed
Database entries
- Structure: 2K77 (N-terminal domain)
- Swiss prot entry: P37571
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: clpC::tet available from the Hamoen] Lab
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal GFP fusions (single copy, also as CFP and YFP variants) available from the Hamoen] Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Your additional remarks
References
Douglas J Kojetin, Patrick D McLaughlin, Richele J Thompson, David Dubnau, Peter Prepiak, Mark Rance, John Cavanagh
Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J Mol Biol: 2009, 387(3);639-52
[PubMed:19361434]
[WorldCat.org]
[DOI]
(I p)
Janine Kirstein, Henrik Strahl, Noël Molière, Leendert W Hamoen, Kürşad Turgay
Localization of general and regulatory proteolysis in Bacillus subtilis cells.
Mol Microbiol: 2008, 70(3);682-94
[PubMed:18786145]
[WorldCat.org]
[DOI]
(I p)
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Holger Kock, Ulf Gerth, Michael Hecker
MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis.
Mol Microbiol: 2004, 51(4);1087-102
[PubMed:14763982]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed