Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' undecaprenyl pyrophosphate phosphatase, bacitracin resistance <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''bcrC'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ywoA '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || undecaprenyl pyrophosphate phosphatase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || resistance to | + | |style="background:#ABCDEF;" align="center"|'''Function''' || resistance to bacitracin and oxidative stress |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 9.455 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 579 bp, 193 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[nrgB]]'', ''[[ywnJ]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15670]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:ywoA_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU36530 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 41: | Line 41: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' | + | * '''DBTBS entry:''' no entry |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12488] |
=== Additional information=== | === Additional information=== | ||
| Line 54: | Line 54: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' bcrC/ybjG/ywoA family (according to Swiss-Prot) |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 72: | Line 72: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' | + | * '''Localization:''' cell membrane (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 78: | Line 78: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P94571 P94571] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU36530] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''bcrC'' |
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' [[SigM]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17434969 PubMed], [[SigI]] [http://www.ncbi.nlm.nih.gov/sites/entrez/18156261 PubMed] |
| − | * '''Regulation:''' | + | * '''Regulation:''' expression activated by glucose (3.3 fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 118: | Line 118: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>12850135 17434969 18156261, </pubmed> |
| − | # | + | # Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in ''Bacillus subtilis'': regulation of the central metabolic pathways. ''Metab Eng.'' '''5:''' 133-149 [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] |
| + | # Jervis et al. 2007. SigM-responsive genes of ''Bacillus subtilis'' and their promoters. J. Bacteriol. 189: 4534-4538. [http://www.ncbi.nlm.nih.gov/sites/entrez/17434969 PubMed] | ||
| + | # Tseng, C. L., and Shaw, G. C. (2008) Genetic evidence for the actin homolog gene ''mreBH'' and the bacitracin resistance gene ''bcrC'' as targets of the alternative [[sigma factor]] SigI of ''Bacillus subtilis''. ''J. Bacteriol.'' '''190:''' 1561-1567. [http://www.ncbi.nlm.nih.gov/sites/entrez/18156261 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 13:09, 8 June 2009
- Description: undecaprenyl pyrophosphate phosphatase, bacitracin resistance
| Gene name | bcrC |
| Synonyms | ywoA |
| Essential | no |
| Product | undecaprenyl pyrophosphate phosphatase |
| Function | resistance to bacitracin and oxidative stress |
| MW, pI | 21 kDa, 9.455 |
| Gene length, protein length | 579 bp, 193 aa |
| Immediate neighbours | nrgB, ywnJ |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 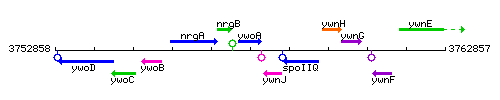
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU36530
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bcrC/ybjG/ywoA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P94571
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon: bcrC
- Regulation: expression activated by glucose (3.3 fold) PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Chi-Ling Tseng, Gwo-Chyuan Shaw
Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis.
J Bacteriol: 2008, 190(5);1561-7
[PubMed:18156261]
[WorldCat.org]
[DOI]
(I p)
Adrian J Jervis, Penny D Thackray, Chris W Houston, Malcolm J Horsburgh, Anne Moir
SigM-responsive genes of Bacillus subtilis and their promoters.
J Bacteriol: 2007, 189(12);4534-8
[PubMed:17434969]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
- Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 5: 133-149 PubMed
- Jervis et al. 2007. SigM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 189: 4534-4538. PubMed
- Tseng, C. L., and Shaw, G. C. (2008) Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J. Bacteriol. 190: 1561-1567. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed