Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' catabolic glutamate dehydrogenase induced by arginine, ornithine or proline, subject to carbon catabolite repression <br/><br/> |
| − | + | {| align="right" border="1" cellpadding="2" | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"|'''Gene name''' | |
| − | + | |''rocG'' | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"| '''Product''' || glutamate dehydrogenase (major) | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"|'''Function''' || arginine utilization, controls the activity of GltC | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 46.2 kDa, 6.28 | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1272 bp, 424 amino acids | |
| − | + | |- | |
| − | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yweA]]'', ''[[rocA]]'' | |
| − | + | |- | |
| − | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/rocG_nucleotide.txt Gene sequence (+200bp) corrected ]''' | |
| − | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/rocG_protein.txt Protein sequence]''' | |
| − | + | |- | |
| − | + | |colspan="2" | '''Genetic context''' <br/> [[Image:rocG_context.gif]] | |
| − | + | |- | |
| − | + | |} | |
| − | + | __TOC__ | |
| − | + | <br/><br/> | |
| − | |||
| − | + | =The gene= | |
| − | + | === Basic information === | |
| − | + | * '''Coordinates:''' 3879765 - 3881036 | |
| − | + | ===Phenotypes of a mutant === | |
| − | + | Poor growth on complex media such as LB. No growth in minimal media with arginine as the only carbon source. Rapid accumulation of suppressor mutants ([[gudB |''gudB1'']]) | |
| − | + | === Database entries === | |
| − | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/rocG.html] | |
| + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10621] | ||
| − | + | === Additional information=== | |
| − | + | =The protein= | |
| − | + | === Basic information/ Evolution === | |
| − | + | * '''Catalyzed reaction/ biological activity:''' L-glutamate + H(2)O + NAD(+) = 2-oxoglutarate + NH(3) + NADH, controls the activity of the [[GltC]] transcription activator [http://www.ncbi.nlm.nih.gov/sites/entrez/17608797 PubMed] | |
| − | + | * '''Protein family:''' Glu/Leu/Phe/Val dehydrogenases family | |
| − | + | * '''Paralogous protein(s):''' [[GudB]] | |
| − | + | === Extended information on the protein === | |
| − | + | * '''Kinetic information:''' | |
| − | + | * '''Domains:''' | |
| − | + | * '''Modification:''' | |
| − | + | * '''Cofactor(s):''' | |
| − | + | * '''Effectors of protein activity:''' | |
| − | + | * '''Interactions:''' RocG-[[GltC]], this interaction prevents transcription activation of the ''gltAB'' operon by GltC [http://www.ncbi.nlm.nih.gov/sites/entrez/17608797 PubMed] | |
| − | + | * '''Localization:''' | |
| − | + | === Database entries === | |
| − | + | * '''Structure:''' | |
| − | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P39633] | |
| − | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU37790] | |
| − | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/1.4.1.2] | |
| − | + | === Additional information=== | |
| − | |||
| − | + | =Expression and regulation= | |
| − | + | * '''Operon:''' ''rocG'' | |
| − | + | * '''Sigma factor:''' [[SigL]] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+10468601 PubMed] | |
| − | + | * '''Regulation:''' induced by arginine ([[RocR]], [[AhrC]]), ornithine or proline, subject to carbon catabolite repression ([[CcpA]]) | |
| − | + | * '''Regulatory mechanism:''' [[RocR]]: transcription activation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+12634342 PubMed][http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+10468601 PubMed]; [[AhrC]]: transcription activation ; [[CcpA]]: transcription repression | |
| − | + | * '''Additional information:''' | |
| + | Activation by RocR requires binding of RocG to a downstream element [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+12634342 PubMed] | ||
| − | + | =Biological materials = | |
| − | |||
| − | |||
| − | + | * '''Mutant:''' GP747 (spc), GP726 (aphA3), available in [[Stülke]] lab | |
| − | + | * '''Expression vector:''' pGP902 (in [[pGP172]], N-terminal Strep-tag), a series of ''rocG'' variants is also available in [[pGP172]], available in [[Stülke]] lab | |
| + | |||
| + | * '''lacZ fusion:''' | ||
| − | + | * '''GFP fusion:''' | |
| − | + | * '''two-hybrid system:''' ''B. pertussis'' adenylate cyclase-based bacterial two hybrid system ([[BACTH]]), available in [[Stülke]] lab | |
| − | + | * '''Antibody:''' available in [[Stülke]] lab | |
| − | + | =Labs working on this gene/protein= | |
| − | + | [[Linc Sonenshein|Linc Sonenshein]], Tufts University, Boston, MA, USA [http://www.tufts.edu/sackler/microbiology/faculty/sonenshein/index.html Homepage] | |
| + | |||
| + | [[Stülke|Jörg Stülke]], University of Göttingen, Germany | ||
| + | [http://wwwuser.gwdg.de/~genmibio/stuelke.html Homepage] | ||
| + | |||
| + | =Your additional remarks= | ||
| + | |||
| + | =References= | ||
| + | |||
| + | # Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of ''Bacillus subtilis'' mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. [http://www.ncbi.nlm.nih.gov/sites/entrez/17183217 PubMed] | ||
| + | # Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in ''Bacillus subtilis'': The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. [http://www.ncbi.nlm.nih.gov/sites/entrez/17608797 PubMed] | ||
| + | # Commichau, F. M., Gunka, K., Landmann, J. J. & Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: Gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J. Bacteriol. 190: 3557-3564. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+18326565 PubMed] | ||
| + | # Herzberg, C., Flórez Weidinger, L. A., Dörrbecker, B., Hübner, S., Stülke, J. & Commichau, F. M. (2007) SPINE: A method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics 7: 4032-4035. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17994626 PubMed] | ||
| + | # Ali, N. O., J. Jeusset, E. Larquet, E. le Cam, B. Belitsky, A. L. Sonenshein, T. Msadek, and M. Débarbouillé. 2003. Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149: 739-750. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+12634342 PubMed] | ||
| + | # Belitsky BR, Sonenshein AL (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298-6305 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+9829940 PubMed] | ||
| + | # Belitsky BR, Sonenshein, AL: An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci USA 1999, 96:10290-10295. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+10468601 PubMed] | ||
| + | # Belitsky BR, Sonenshein, AL: CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J Bacteriol 2004, 186:3392-3398. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+15150224 PubMed] | ||
| + | # Belitsky BR, Sonenshein AL (2004) Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186:3399-3407 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+15150225 PubMed] | ||
| + | # Khan, M. I., K. Ito, H. Kim, H. Ashida, T. Ishikawa, H. Shibata, and Y. Sawa. 2005. Molecular properties and enhancement of thermostability by random mutagenesis of glutamate dehydrogenase from Bacillus subtilis. Biosci. Biotechnol. Biochem. 69: 1861-1870. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+16244435 PubMed] | ||
| + | # Stillman TJ, Baker PJ, Britton KL, Rice DW Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis. J Mol Biol 1993, 234:1131-1139. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+8263917 PubMed] | ||
Revision as of 15:21, 2 February 2009
- Description: catabolic glutamate dehydrogenase induced by arginine, ornithine or proline, subject to carbon catabolite repression
| Gene name | rocG |
| Synonyms | |
| Essential | no |
| Product | glutamate dehydrogenase (major) |
| Function | arginine utilization, controls the activity of GltC |
| MW, pI | 46.2 kDa, 6.28 |
| Gene length, protein length | 1272 bp, 424 amino acids |
| Immediate neighbours | yweA, rocA |
| Gene sequence (+200bp) corrected | Protein sequence |
Genetic context 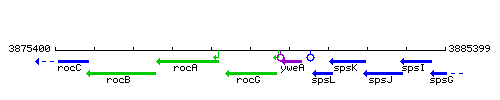
| |
Contents
The gene
Basic information
- Coordinates: 3879765 - 3881036
Phenotypes of a mutant
Poor growth on complex media such as LB. No growth in minimal media with arginine as the only carbon source. Rapid accumulation of suppressor mutants (gudB1)
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate + H(2)O + NAD(+) = 2-oxoglutarate + NH(3) + NADH, controls the activity of the GltC transcription activator PubMed
- Protein family: Glu/Leu/Phe/Val dehydrogenases family
- Paralogous protein(s): GudB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: RocG-GltC, this interaction prevents transcription activation of the gltAB operon by GltC PubMed
- Localization:
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Operon: rocG
- Regulation: induced by arginine (RocR, AhrC), ornithine or proline, subject to carbon catabolite repression (CcpA)
- Regulatory mechanism: RocR: transcription activation PubMedPubMed; AhrC: transcription activation ; CcpA: transcription repression
- Additional information:
Activation by RocR requires binding of RocG to a downstream element PubMed
Biological materials
- Mutant: GP747 (spc), GP726 (aphA3), available in Stülke lab
- Expression vector: pGP902 (in pGP172, N-terminal Strep-tag), a series of rocG variants is also available in pGP172, available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody: available in Stülke lab
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. PubMed
- Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. PubMed
- Commichau, F. M., Gunka, K., Landmann, J. J. & Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: Gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J. Bacteriol. 190: 3557-3564. PubMed
- Herzberg, C., Flórez Weidinger, L. A., Dörrbecker, B., Hübner, S., Stülke, J. & Commichau, F. M. (2007) SPINE: A method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics 7: 4032-4035. PubMed
- Ali, N. O., J. Jeusset, E. Larquet, E. le Cam, B. Belitsky, A. L. Sonenshein, T. Msadek, and M. Débarbouillé. 2003. Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149: 739-750. PubMed
- Belitsky BR, Sonenshein AL (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298-6305 PubMed
- Belitsky BR, Sonenshein, AL: An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci USA 1999, 96:10290-10295. PubMed
- Belitsky BR, Sonenshein, AL: CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J Bacteriol 2004, 186:3392-3398. PubMed
- Belitsky BR, Sonenshein AL (2004) Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186:3399-3407 PubMed
- Khan, M. I., K. Ito, H. Kim, H. Ashida, T. Ishikawa, H. Shibata, and Y. Sawa. 2005. Molecular properties and enhancement of thermostability by random mutagenesis of glutamate dehydrogenase from Bacillus subtilis. Biosci. Biotechnol. Biochem. 69: 1861-1870. PubMed
- Stillman TJ, Baker PJ, Britton KL, Rice DW Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis. J Mol Biol 1993, 234:1131-1139. PubMed