Difference between revisions of "Sandbox"
Raphael2215 (talk | contribs) |
|||
| Line 25: | Line 25: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[acoL]]'', ''[[sspH]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[acoL]]'', ''[[sspH]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU00100 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU00100 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU00100 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:acoR_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:acoR_context.gif]] | ||
Revision as of 11:26, 13 May 2013
- Achtung: transcriptional activator of the acoA-acoB-acoC-acoL operon
| Gene name | acoR |
| Synonyms | yfjG, yzcB |
| Essential | no |
| Product | transcriptional activator (for SigL-dependent promoter) |
| Function | regulation of acetoin utilization |
| Gene expression levels in SubtiExpress: eno | |
| Interactions involving this protein in SubtInteract: Eno | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 66 kDa, 7.884 |
| Gene length, protein length | 1815 bp, 605 aa |
| Immediate neighbours | acoL, sspH |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 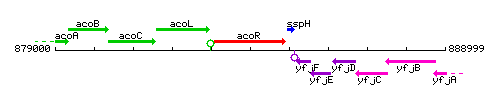
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed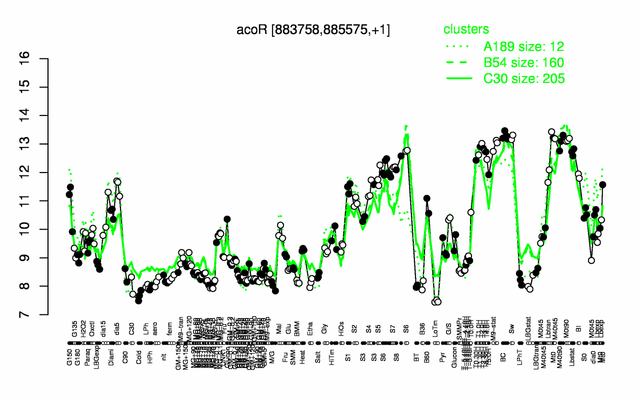
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control
This gene is a member of the following regulons
The AcoR regulon:
The gene
Basic information
- Locus tag: BSU08100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31551
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Michel Debarbouille, Pasteur Institute, Paris, France Homepage
Your additional remarks
References
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
N O Ali, J Bignon, G Rapoport, M Debarbouille
Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis.
J Bacteriol: 2001, 183(8);2497-504
[PubMed:11274109]
[WorldCat.org]
[DOI]
(P p)
M Huang, F B Oppermann-Sanio, A Steinbüchel
Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway.
J Bacteriol: 1999, 181(12);3837-41
[PubMed:10368162]
[WorldCat.org]
[DOI]
(P p)
hello