Difference between revisions of "SacY"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[utilization of specific carbon sources]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| + | {{SubtiWiki category|[[RNA binding regulators]]}}, | ||
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[SacY regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 60: | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 00:43, 9 December 2010
- Description: transcriptional antiterminator for sacB and sacX-sacY (acts at high sucrose concentrations)
| Gene name | sacY |
| Synonyms | ipa-13r, sacS |
| Essential | no |
| Product | transcriptional antiterminator |
| Function | regulation of sucrose utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism, Stress | |
| MW, pI | 32 kDa, 5.883 |
| Gene length, protein length | 840 bp, 280 aa |
| Immediate neighbours | sacX, gspA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 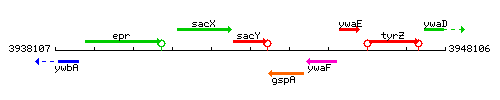
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control, RNA binding regulators, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38420
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binding to the mRNA of sacB and the sacX-sacY operon, causes transcription antitermination (in presence of sucrose)
- Protein family: transcriptional antiterminator bglG family (according to Swiss-Prot) BglG family of antiterminators
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1AUU (RNA-binding domain)
- UniProt: P15401
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: induction by sucrose (at high concentration) PubMed
- Regulatory mechanism:
- induction by SacY-dependent RNA switch (transcriptional antitermination) PubMed
- DegU-P: transcription activation PubMed
- Additional information:
Biological materials
- Mutant: GP425 (cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
P Tortosa, N Declerck, H Dutartre, C Lindner, J Deutscher, D Le Coq
Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY.
Mol Microbiol: 2001, 41(6);1381-93
[PubMed:11580842]
[WorldCat.org]
[DOI]
(P p)
N Declerck, F Vincent, F Hoh, S Aymerich, H van Tilbeurgh
RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT.
J Mol Biol: 1999, 294(2);389-402
[PubMed:10610766]
[WorldCat.org]
[DOI]
(P p)
H van Tilbeurgh, X Manival, S Aymerich, J M Lhoste, C Dumas, M Kochoyan
Crystal structure of a new RNA-binding domain from the antiterminator protein SacY of Bacillus subtilis.
EMBO J: 1997, 16(16);5030-6
[PubMed:9305644]
[WorldCat.org]
[DOI]
(P p)
X Manival, Y Yang, M P Strub, M Kochoyan, M Steinmetz, S Aymerich
From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins.
EMBO J: 1997, 16(16);5019-29
[PubMed:9305643]
[WorldCat.org]
[DOI]
(P p)
P Tortosa, S Aymerich, C Lindner, M H Saier, J Reizer, D Le Coq
Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system.
J Biol Chem: 1997, 272(27);17230-7
[PubMed:9202047]
[WorldCat.org]
[DOI]
(P p)
M Arnaud, M Débarbouillé, G Rapoport, M H Saier, J Reizer
In vitro reconstitution of transcriptional antitermination by the SacT and SacY proteins of Bacillus subtilis.
J Biol Chem: 1996, 271(31);18966-72
[PubMed:8702561]
[WorldCat.org]
[DOI]
(P p)
P Tortosa, D Le Coq
A ribonucleic antiterminator sequence (RAT) and a distant palindrome are both involved in sucrose induction of the Bacillus subtilis sacXY regulatory operon.
Microbiology (Reading): 1995, 141 ( Pt 11);2921-7
[PubMed:8535520]
[WorldCat.org]
[DOI]
(P p)
S Aymerich, M Steinmetz
Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family.
Proc Natl Acad Sci U S A: 1992, 89(21);10410-4
[PubMed:1279678]
[WorldCat.org]
[DOI]
(P p)
A M Crutz, M Steinmetz
Transcription of the Bacillus subtilis sacX and sacY genes, encoding regulators of sucrose metabolism, is both inducible by sucrose and controlled by the DegS-DegU signalling system.
J Bacteriol: 1992, 174(19);6087-95
[PubMed:1400159]
[WorldCat.org]
[DOI]
(P p)
M M Zukowski, L Miller, P Cosgwell, K Chen, S Aymerich, M Steinmetz
Nucleotide sequence of the sacS locus of Bacillus subtilis reveals the presence of two regulatory genes.
Gene: 1990, 90(1);153-5
[PubMed:2116367]
[WorldCat.org]
[DOI]
(P p)
A M Crutz, M Steinmetz, S Aymerich, R Richter, D Le Coq
Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system.
J Bacteriol: 1990, 172(2);1043-50
[PubMed:2105292]
[WorldCat.org]
[DOI]
(P p)