Difference between revisions of "RsbV"
| Line 63: | Line 63: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU04710&redirect=T BSU04710] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/rsbRSTUVW-sigB-rsbX.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/rsbRSTUVW-sigB-rsbX.html] | ||
| Line 104: | Line 105: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU04710&redirect=T BSU04710] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1VC1 1VC1] (homolog from Thermotoga maritima) | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1VC1 1VC1] (homolog from Thermotoga maritima) | ||
Revision as of 13:02, 2 April 2014
| Gene name | rsbV |
| Synonyms | |
| Essential | no |
| Product | anti-anti-SigB |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: rsbV | |
| Interactions involving this protein in SubtInteract: RsbV | |
| Metabolic function and regulation of this protein in SubtiPathways: rsbV | |
| MW, pI | 11 kDa, 4.698 |
| Gene length, protein length | 327 bp, 109 aa |
| Immediate neighbours | rsbU, rsbW |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed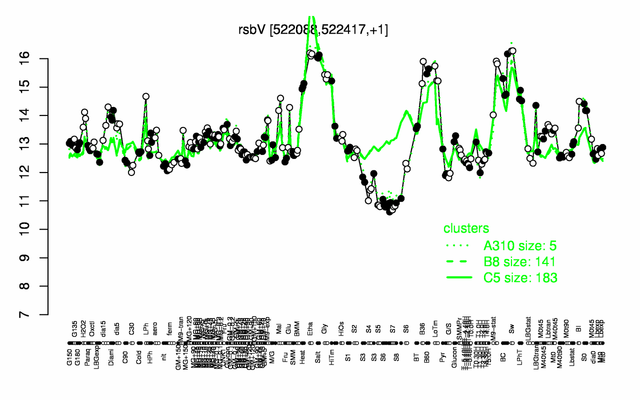
| |
Contents
Categories containing this gene/protein
sigma factors and their control, general stress proteins (controlled by SigB), phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04710
Phenotypes of a mutant
Database entries
- BsubCyc: BSU04710
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: STAS domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU04710
- Structure: 1VC1 (homolog from Thermotoga maritima)
- UniProt: P17903
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- CcpA: transcription repression PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Locke JC, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB Stochastic pulse regulation in bacterial stress response. Science. 2011 334:366-369. PubMed:21979936
Soo-Keun Choi, Milton H Saier
Transcriptional Regulation of the rsbV Promoter Controlling Stress Responses to Ethanol, Carbon Limitation, and Phosphorous Limitation in Bacillus subtilis.
Int J Microbiol: 2010, 2010;263410
[PubMed:20454630]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Gudrun Holtmann, Matthias Brigulla, Leif Steil, Alexandra Schütz, Karsta Barnekow, Uwe Völker, Erhard Bremer
RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature.
J Bacteriol: 2004, 186(18);6150-8
[PubMed:15342585]
[WorldCat.org]
[DOI]
(P p)
Emmanuel Guedon, Charles M Moore, Qiang Que, Tao Wang, Rick W Ye, John D Helmann
The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons.
Mol Microbiol: 2003, 49(6);1477-91
[PubMed:12950915]
[WorldCat.org]
[DOI]
(P p)
Matthias Brigulla, Tamara Hoffmann, Andrea Krisp, Andrea Völker, Erhard Bremer, Uwe Völker
Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation.
J Bacteriol: 2003, 185(15);4305-14
[PubMed:12867438]
[WorldCat.org]
[DOI]
(P p)
Olivier Delumeau, Richard J Lewis, Michael D Yudkin
Protein-protein interactions that regulate the energy stress activation of sigma(B) in Bacillus subtilis.
J Bacteriol: 2002, 184(20);5583-9
[PubMed:12270815]
[WorldCat.org]
[DOI]
(P p)
M S Brody, K Vijay, C W Price
Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis.
J Bacteriol: 2001, 183(21);6422-8
[PubMed:11591687]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
K Vijay, M S Brody, E Fredlund, C W Price
A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis.
Mol Microbiol: 2000, 35(1);180-8
[PubMed:10632888]
[WorldCat.org]
[DOI]
(P p)
N Smirnova, J Scott, U Voelker, W G Haldenwang
Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulator RsbX.
J Bacteriol: 1998, 180(14);3671-80
[PubMed:9658013]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
The yeast two-hybrid system detects interactions between Bacillus subtilis sigmaB regulators.
J Bacteriol: 1996, 178(23);7020-3
[PubMed:8955331]
[WorldCat.org]
[DOI]
(P p)
X Yang, C M Kang, M S Brody, C W Price
Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor.
Genes Dev: 1996, 10(18);2265-75
[PubMed:8824586]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities.
J Bacteriol: 1996, 178(18);5456-63
[PubMed:8808936]
[WorldCat.org]
[DOI]
(P p)
S Alper, A Dufour, D A Garsin, L Duncan, R Losick
Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis.
J Mol Biol: 1996, 260(2);165-77
[PubMed:8764398]
[WorldCat.org]
[DOI]
(P p)
C M Kang, M S Brody, S Akbar, X Yang, C W Price
Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress.
J Bacteriol: 1996, 178(13);3846-53
[PubMed:8682789]
[WorldCat.org]
[DOI]
(P p)
A Dufour, U Voelker, A Voelker, W G Haldenwang
Relative levels and fractionation properties of Bacillus subtilis σ(B) and its regulators during balanced growth and stress.
J Bacteriol: 1996, 178(13);3701-9 sigma
[PubMed:8682769]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, B Maul, M Hecker, A Dufour, W G Haldenwang
Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses.
J Bacteriol: 1995, 177(13);3771-80
[PubMed:7601843]
[WorldCat.org]
[DOI]
(P p)
A A Wise, C W Price
Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals.
J Bacteriol: 1995, 177(1);123-33
[PubMed:8002610]
[WorldCat.org]
[DOI]
(P p)
A Dufour, W G Haldenwang
Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV).
J Bacteriol: 1994, 176(7);1813-20
[PubMed:8144446]
[WorldCat.org]
[DOI]
(P p)