Difference between revisions of "RsbR"
| Line 76: | Line 76: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3VY9 3VY9] (complete stressosome) | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2BNL 2BNL] (N-terminal domain), [http://www.rcsb.org/pdb/explore.do?structureId=3VY9 3VY9] (complete stressosome) |
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P42409 P42409] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P42409 P42409] | ||
Revision as of 21:50, 5 May 2009
- Description: activator of RsbT kinase activity, part of the stressosome
| Gene name | rsbR |
| Synonyms | ycxR, rsbRA |
| Essential | no |
| Product | activator of RsbT kinase activity |
| Function | control of SigB activity |
| MW, pI | 30 kDa, 4.731 |
| Gene length, protein length | 822 bp, 274 aa |
| Immediate neighbours | ndoA, rsbS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 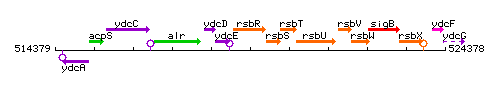
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Swiss prot entry: P42409
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Marles-Wright, J., Grant, T., Delumeau, O., van Duinen, G., Firbank, S. J., Lewis, P. J., Murray, J. W., Newman, J. A., Quin, M. B., Race, P. R., Rohou, A., Tichelaar, W., van Heel, M. & Lewis, R. J. (2008) Molecular architecture of the "stressosome," a signal integration and transduction hub. Science 322: 92-96. PubMed
- Eymann et al. (2007) Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 7: 3509-3526. PubMed
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed