Difference between revisions of "RsbP"
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU34110 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 80: | Line 80: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O07014 O07014] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O07014 O07014] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU34110] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/3.1.3.3 3.1.3.3] | * '''E.C. number:''' [http://www.expasy.org/enzyme/3.1.3.3 3.1.3.3] | ||
Revision as of 14:13, 3 June 2009
- Description: protein serine phosphatase, energy PP2C, dephosphorylates RsbV
| Gene name | rsbP |
| Synonyms | yvfP |
| Essential | no |
| Product | protein serine phosphatase, energy PP2C |
| Function | control of SigB activity |
| MW, pI | 45 kDa, 4.827 |
| Gene length, protein length | 1209 bp, 403 aa |
| Immediate neighbours | rsbQ, yvfO |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 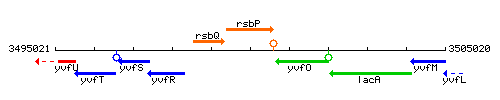
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU34110
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: O-phospho-L(or D)-serine + H2O = L(or D)-serine + phosphate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains: N-terminal PAS domain, central coiled-coil domain, C-terminal PP2C phosphatase domain
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- Swiss prot entry: O07014
- KEGG entry: [2]
- E.C. number: 3.1.3.3
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Chet Price, Davis, USA homepage
Your additional remarks
References
Masatomo Makino, Shinpei Kondo, Tomonori Kaneko, Seiki Baba, Kunio Hirata, Takashi Kumasaka
Expression, crystallization and preliminary crystallographic analysis of the PAS domain of RsbP, a stress-response phosphatase from Bacillus subtilis.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 6);559-61
[PubMed:19478430]
[WorldCat.org]
[DOI]
(I p)
Margaret S Brody, Valley Stewart, Chester W Price
Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis.
Mol Microbiol: 2009, 72(5);1221-34
[PubMed:19432806]
[WorldCat.org]
[DOI]
(I p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed