Difference between revisions of "RpsN"
| Line 45: | Line 45: | ||
* '''Locus tag:''' BSU01290 | * '''Locus tag:''' BSU01290 | ||
| + | |||
| + | [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rpsNA_141757_141942_1 Expression] | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 12:25, 24 January 2012
- Description: ribosomal protein
| Gene name | rpsN |
| Synonyms | |
| Essential | yes PubMed |
| Product | ribosomal protein S14 |
| Function | translation |
| Interactions involving this protein in SubtInteract: RpsN | |
| MW, pI | 7 kDa, 10.914 |
| Gene length, protein length | 183 bp, 61 aa |
| Immediate neighbours | rplE, rpsH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 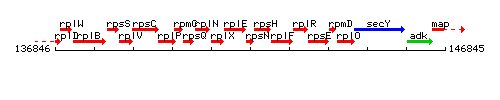
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01290
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein S14P family (according to Swiss-Prot) Zinc-binding S14P subfamily (according to Swiss-Prot)
- Paralogous protein(s): YhzA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P12878
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon: rpsJ-rplC-rplD-rplW-rplB-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-rplX-rplE-rpsN-rpsH-rplF-rplR-rpsE-rpmD-rplO PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Hideaki Nanamiya, Fujio Kawamura
Towards an elucidation of the roles of the ribosome during different growth phases in Bacillus subtilis.
Biosci Biotechnol Biochem: 2010, 74(3);451-61
[PubMed:20208344]
[WorldCat.org]
[DOI]
(I p)
Matthew A Lauber, William E Running, James P Reilly
B. subtilis ribosomal proteins: structural homology and post-translational modifications.
J Proteome Res: 2009, 8(9);4193-206
[PubMed:19653700]
[WorldCat.org]
[DOI]
(P p)
Scott E Gabriel, John D Helmann
Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions.
J Bacteriol: 2009, 191(19);6116-22
[PubMed:19648245]
[WorldCat.org]
[DOI]
(I p)
Yousuke Natori, Hideaki Nanamiya, Genki Akanuma, Saori Kosono, Toshiaki Kudo, Kozo Ochi, Fujio Kawamura
A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis.
Mol Microbiol: 2007, 63(1);294-307
[PubMed:17163968]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
X Li, L Lindahl, Y Sha, J M Zengel
Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster.
J Bacteriol: 1997, 179(22);7046-54
[PubMed:9371452]
[WorldCat.org]
[DOI]
(P p)
J W Suh, S A Boylan, S H Oh, C W Price
Genetic and transcriptional organization of the Bacillus subtilis spc-alpha region.
Gene: 1996, 169(1);17-23
[PubMed:8635744]
[WorldCat.org]
[DOI]
(P p)