Difference between revisions of "RpsF"
| Line 108: | Line 108: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rpsF_4199445_4199732_-1 rpsF] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rpsF_4199445_4199732_-1 rpsF] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 135: | Line 135: | ||
=References= | =References= | ||
| − | + | <pubmed>19653700 14762004 11948165 11948146 23002217</pubmed> | |
| − | <pubmed>19653700 14762004 11948165 11948146</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:45, 19 June 2013
- Description: ribosomal protein
| Gene name | rpsF |
| Synonyms | |
| Essential | no PubMed |
| Product | ribosomal protein S6 (BS9) |
| Function | translation |
| Gene expression levels in SubtiExpress: rpsF | |
| Interactions involving this protein in SubtInteract: RpsF | |
| MW, pI | 10 kDa, 4.993 |
| Gene length, protein length | 285 bp, 95 aa |
| Immediate neighbours | ssbA, yyaF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 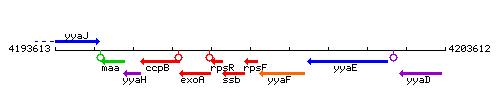
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed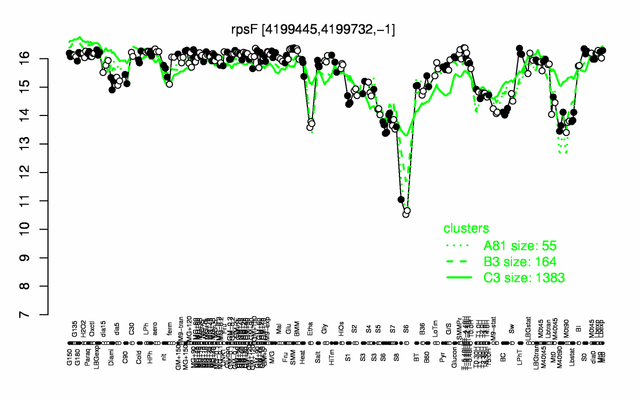
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
ComK regulon, stringent response
The gene
Basic information
- Locus tag: BSU40910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein S6P family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3R3T (from Bacillus anthracis, 71% identity, 97% similarity)
- UniProt: P21468
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Genki Akanuma, Hideaki Nanamiya, Yousuke Natori, Koichi Yano, Shota Suzuki, Shuya Omata, Morio Ishizuka, Yasuhiko Sekine, Fujio Kawamura
Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation.
J Bacteriol: 2012, 194(22);6282-91
[PubMed:23002217]
[WorldCat.org]
[DOI]
(I p)
Matthew A Lauber, William E Running, James P Reilly
B. subtilis ribosomal proteins: structural homology and post-translational modifications.
J Proteome Res: 2009, 8(9);4193-206
[PubMed:19653700]
[WorldCat.org]
[DOI]
(P p)
Cordula Lindner, Reindert Nijland, Mariska van Hartskamp, Sierd Bron, Leendert W Hamoen, Oscar P Kuipers
Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein.
J Bacteriol: 2004, 186(4);1097-105
[PubMed:14762004]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
Mitsuo Ogura, Hirotake Yamaguchi, Kazuo Kobayashi, Naotake Ogasawara, Yasutaro Fujita, Teruo Tanaka
Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK.
J Bacteriol: 2002, 184(9);2344-51
[PubMed:11948146]
[WorldCat.org]
[DOI]
(P p)