Difference between revisions of "RpoB"
(→Biological materials) |
(→Biological materials) |
||
| Line 154: | Line 154: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| + | ** GP2047 (aphA3) based on [[pAC7]], available in [[Jörg Stülke]]'s lab | ||
| + | ** GP2052 (cat) based on [[pAC5]], available in [[Jörg Stülke]]'s lab | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
Revision as of 07:55, 17 July 2015
- Description: RNA polymerase beta subunit
| Gene name | rpoB |
| Synonyms | |
| Essential | yes PubMed |
| Product | RNA polymerase beta subunit |
| Function | transcription |
| Gene expression levels in SubtiExpress: rpoB | |
| Interactions involving this protein in SubtInteract: RpoB | |
| MW, pI | 133 kDa, 4.731 |
| Gene length, protein length | 3579 bp, 1193 aa |
| Immediate neighbours | ybxB, rpoC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 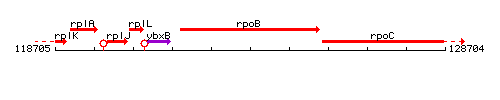
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed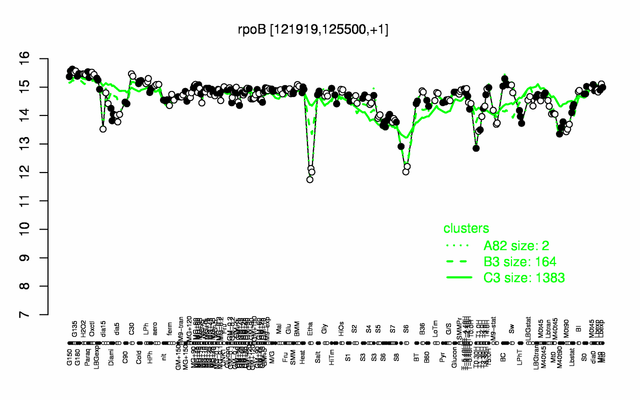
| |
Contents
Categories containing this gene/protein
transcription, essential genes, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01070
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01070
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
- mutations in mtrB, sigB, rpoB, and rpoC allow B. subtilis to grow with 4-fluorotryptophan rather than with tryptophan as a canonical amino acid of the genetic code PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1) (according to Swiss-Prot)
- Protein family: RNA polymerase beta chain family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on (Arg-312 OR Arg-313), Arg-539, (Arg-693 OR Arg-694), Arg-827, and Arg-1106 PubMed
- Effectors of protein activity:
- Interactions:
- RpoA-RpoB-RpoC PubMed, NusA-RpoB PubMed
- SigA-(RpoB-RpoC) PubMed, SigB-(RpoB-RpoC)
- SigD-(RpoB-RpoC), SigE-(RpoB-RpoC)
- SigF-(RpoB-RpoC), SigG-(RpoB-RpoC)
- SigH-(RpoB-RpoC), SigI-(RpoB-RpoC)
- SigK-(RpoB-RpoC), SigL-(RpoB-RpoC)
- SigM-(RpoB-RpoC), SigV-(RpoB-RpoC)
- SigW-(RpoB-RpoC), SigX-(RpoB-RpoC)
- SigY-(RpoB-RpoC), SigZ-(RpoB-RpoC)
- Xpf-(RpoB-RpoC), YlaC-(RpoB-RpoC)
- YvrI-RpoB PubMed,
- Mfd-RpoB PubMed
- RpoB-RpoE (demonstrated for S. aureus) PubMed
- Localization: membrane associated PubMed
Database entries
- BsubCyc: BSU01070
- Structure:
- UniProt: P37870
- KEGG entry: [3]
- E.C. number: 2.7.7.6
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information:
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 3434 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 10168 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 1411 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 800 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 802 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GP2047 (aphA3) based on pAC7, available in Jörg Stülke's lab
- GP2052 (cat) based on pAC5, available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Fabian Commichau's lab
- FLAG-tag construct:
- GP2043 rpoB-3xFLAG ermC (based on pGP1087), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications