Difference between revisions of "RplL"
(→Extended information on the protein) |
(→Expression and regulation) |
||
| Line 111: | Line 111: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[rplJ]]-[[rplL]]'' {{PubMed|26101249}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rplL_120607_120978_1 rplL] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rplL_120607_120978_1 rplL] {{PubMed|22383849}} | ||
| Line 119: | Line 119: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
** [[RelA]] dependent downregulation (Class I) during stringent response {{PubMed|11948165}} | ** [[RelA]] dependent downregulation (Class I) during stringent response {{PubMed|11948165}} | ||
| + | ** termination/ antitermination ([[rplJ|L10]]-[[rplL|L12]]<sub>4</sub> complex) {{PubMed|26101249}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[rplJ|L10]]-[[rplL|L12]]<sub>4</sub> complex: transcription termination (attenuation) at excess of the complex (autorepression) {{PubMed|26101249}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
Revision as of 09:25, 23 July 2015
- Description: ribosomal protein
| Gene name | rplL |
| Synonyms | |
| Essential | yes PubMed |
| Product | ribosomal protein L12 (BL9) |
| Function | translation |
| Gene expression levels in SubtiExpress: rplL | |
| Interactions involving this protein in SubtInteract: RplL | |
| MW, pI | 12 kDa, 4.355 |
| Gene length, protein length | 369 bp, 123 aa |
| Immediate neighbours | rplJ, ybxB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 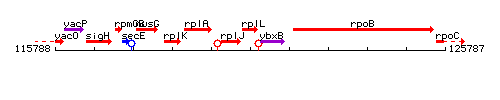
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed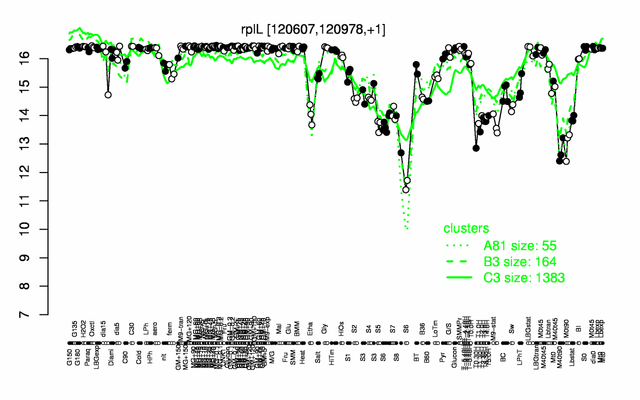
| |
Contents
Categories containing this gene/protein
translation, essential genes, membrane proteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01050
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01050
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein L12P family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: membrane associated PubMed
Database entries
- BsubCyc: BSU01050
- Structure:
- UniProt: P02394
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 24809 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 142727 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 20697 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 11499 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 19060 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Helen Yakhnin, Alexander V Yakhnin, Paul Babitzke
Ribosomal protein L10(L12)4 autoregulates expression of the Bacillus subtilis rplJL operon by a transcription attenuation mechanism.
Nucleic Acids Res: 2015, 43(14);7032-43
[PubMed:26101249]
[WorldCat.org]
[DOI]
(I p)
Yun Chen, Shu Feng, Veerendra Kumar, Rya Ero, Yong-Gui Gao
Structure of EF-G-ribosome complex in a pretranslocation state.
Nat Struct Mol Biol: 2013, 20(9);1077-84
[PubMed:23912278]
[WorldCat.org]
[DOI]
(I p)
Genki Akanuma, Hideaki Nanamiya, Yousuke Natori, Koichi Yano, Shota Suzuki, Shuya Omata, Morio Ishizuka, Yasuhiko Sekine, Fujio Kawamura
Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation.
J Bacteriol: 2012, 194(22);6282-91
[PubMed:23002217]
[WorldCat.org]
[DOI]
(I p)
Matthew A Lauber, William E Running, James P Reilly
B. subtilis ribosomal proteins: structural homology and post-translational modifications.
J Proteome Res: 2009, 8(9);4193-206
[PubMed:19653700]
[WorldCat.org]
[DOI]
(P p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
James R Iben, David E Draper
Specific interactions of the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11.
Biochemistry: 2008, 47(9);2721-31
[PubMed:18247578]
[WorldCat.org]
[DOI]
(P p)
Catherine Wicker-Planquart, Anne-Emmanuelle Foucher, Mathilde Louwagie, Robert A Britton, Jean-Michel Jault
Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes.
J Bacteriol: 2008, 190(2);681-90
[PubMed:17981968]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
K J Boor, M L Duncan, C W Price
Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase.
J Biol Chem: 1995, 270(35);20329-36
[PubMed:7657605]
[WorldCat.org]
[DOI]
(P p)