Difference between revisions of "RocR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcriptional activator of arginine utilization operons <br/><br/> | + | * '''Description:''' [[transcription factors activating transcription at SigL-dependent promoters |transcriptional activator]] of arginine utilization operons <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional | + | |style="background:#ABCDEF;" align="center"| '''Product''' || [[transcription factors activating transcription at SigL-dependent promoters |transcriptional activator]] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || transcriptional activator of arginine utilization operons | |style="background:#ABCDEF;" align="center"|'''Function''' || transcriptional activator of arginine utilization operons | ||
Revision as of 17:51, 27 November 2009
- Description: transcriptional activator of arginine utilization operons
| Gene name | rocR |
| Synonyms | |
| Essential | no |
| Product | transcriptional activator |
| Function | transcriptional activator of arginine utilization operons |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 52.6 kDa, 6.08 |
| Gene length, protein length | 1383 bp, 461 amino acids |
| Immediate neighbours | rocD, yyxA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 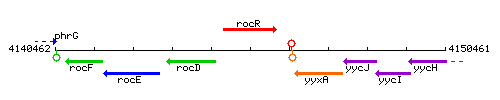
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU40350
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activation at SigL-dependent promoters of rocABC, rocDEF, and rocG
- Protein family: Sigma-54 interacting transcription activator
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Sigma-54 factor interaction domain (143–372)
- 2x nucleotid-binding domain (171–178),(233–242)
- HTH motif (434–453)
- Modification:
- Cofactor(s): ornithine or citrulline are required for RocR-dependent transcription activation PubMed
- Effectors of protein activity:
- Interactions: RocR-SigL
- Localization:
Database entries
- Structure:
- UniProt: P38022
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: rocR
- Regulatory mechanism: autorepression
- Additional information:
Biological materials
- Mutant: QB5533 (aphA3), from Michel Debarbouille, available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Michel Debarbouille, Pasteur Institute, Paris, France Homepage
Your additional remarks
References
Naima Ould Ali, Josette Jeusset, Eric Larquet, Eric Le Cam, Boris Belitsky, Abraham L Sonenshein, Tarek Msadek, Michel Débarbouillé
Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis.
Microbiology (Reading): 2003, 149(Pt 3);739-750
[PubMed:12634342]
[WorldCat.org]
[DOI]
(P p)
S Calogero, R Gardan, P Glaser, J Schweizer, G Rapoport, M Debarbouille
RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators.
J Bacteriol: 1994, 176(5);1234-41
[PubMed:8113162]
[WorldCat.org]
[DOI]
(P p)