Difference between revisions of "RocR"
(→References) |
|||
| Line 131: | Line 131: | ||
=References= | =References= | ||
| − | <pubmed>12634342 8113162,9194709 | + | <pubmed>12634342 8113162,9194709 21965396 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:52, 31 October 2011
- Description: transcriptional activator of arginine utilization operons
| Gene name | rocR |
| Synonyms | |
| Essential | no |
| Product | transcriptional activator |
| Function | transcriptional activator of arginine utilization operons |
| Interactions involving this protein in SubtInteract: RocR | |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 52.6 kDa, 6.08 |
| Gene length, protein length | 1383 bp, 461 amino acids |
| Immediate neighbours | rocD, yyxA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 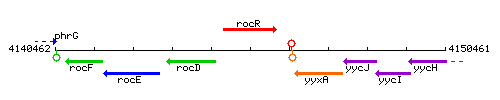
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of amino acids, transcription factors and their control
This gene is a member of the following regulons
The RocR regulon
The gene
Basic information
- Locus tag: BSU40350
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activation at SigL-dependent promoters of rocABC, rocDEF, and rocG
- Protein family: Sigma-54 interacting transcription activator
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Sigma-54 factor interaction domain (143–372)
- 2x nucleotid-binding domain (171–178),(233–242)
- HTH motif (434–453)
- Modification:
- Cofactor(s): ornithine or citrulline are required for RocR-dependent transcription activation PubMed
- Effectors of protein activity:
- Interactions: RocR-SigL
- Localization:
Database entries
- Structure:
- UniProt: P38022
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: rocR (according to DBTBS)
- Regulatory mechanism: autorepression
- Additional information:
Biological materials
- Mutant: QB5533 (aphA3), from Michel Debarbouille, available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Michel Debarbouille, Pasteur Institute, Paris, France Homepage
Your additional remarks
References
Kenji Manabe, Yasushi Kageyama, Takuya Morimoto, Tadahiro Ozawa, Kazuhisa Sawada, Keiji Endo, Masatoshi Tohata, Katsutoshi Ara, Katsuya Ozaki, Naotake Ogasawara
Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874.
Appl Environ Microbiol: 2011, 77(23);8370-81
[PubMed:21965396]
[WorldCat.org]
[DOI]
(I p)
Naima Ould Ali, Josette Jeusset, Eric Larquet, Eric Le Cam, Boris Belitsky, Abraham L Sonenshein, Tarek Msadek, Michel Débarbouillé
Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis.
Microbiology (Reading): 2003, 149(Pt 3);739-750
[PubMed:12634342]
[WorldCat.org]
[DOI]
(P p)
R Gardan, G Rapoport, M Débarbouillé
Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis.
Mol Microbiol: 1997, 24(4);825-37
[PubMed:9194709]
[WorldCat.org]
[DOI]
(P p)
S Calogero, R Gardan, P Glaser, J Schweizer, G Rapoport, M Debarbouille
RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators.
J Bacteriol: 1994, 176(5);1234-41
[PubMed:8113162]
[WorldCat.org]
[DOI]
(P p)