Difference between revisions of "RocF"
(→Extended information on the protein) |
|||
| Line 103: | Line 103: | ||
* '''Operon:''' ''[[rocD]]-[[rocE]]-[[rocF]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/7540694 PubMed] | * '''Operon:''' ''[[rocD]]-[[rocE]]-[[rocF]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/7540694 PubMed] | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=argI_4141711_4142601_-1 rocF] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigL]] [http://www.ncbi.nlm.nih.gov/sites/entrez/7540694 PubMed] | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 10:24, 17 April 2012
- Description: arginase
| Gene name | rocF |
| Synonyms | |
| Essential | no |
| Product | arginase |
| Function | arginine utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 32 kDa, 4.932 |
| Gene length, protein length | 888 bp, 296 aa |
| Immediate neighbours | phrG, rocE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 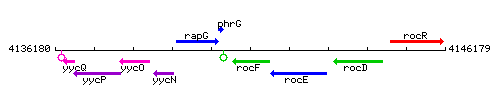
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
AhrC regulon, CodY regulon, RocR regulon, SigL regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU40320
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-arginine + H2O = L-ornithine + urea (according to Swiss-Prot)
- Protein family: arginase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on Ser-68 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 2EF5 (from Thermus thermophilus, 46% identity, 63% similarity)
- UniProt: P39138
- KEGG entry: [3]
- E.C. number: 3.5.3.1
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP655, aphA3, available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
R Gardan, G Rapoport, M Débarbouillé
Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis.
J Mol Biol: 1995, 249(5);843-56
[PubMed:7540694]
[WorldCat.org]
[DOI]
(P p)