Difference between revisions of "Rny"
(→References) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' RNase Y, endoribonuclease, | + | * '''Description:''' RNase Y, 5' end sensitive endoribonuclease, involved in the degradation/processing of mRNA<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || RNase Y | |style="background:#ABCDEF;" align="center"| '''Product''' || RNase Y | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || required for the processing <br/>of the ''[[gapA]]'' operon mRNA | + | |style="background:#ABCDEF;" align="center"|'''Function''' || Initiates S-box riboswitch RNA turnover, required for the processing <br/>of the ''[[gapA]]'' operon mRNA, depletion of RNase Y increases bulk mRNA stability. |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Regulatory function of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Regulatory function of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
| Line 57: | Line 57: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' RNase Y cleaves S-box riboswitch RNAs ''in vivo'' and ''in vitro''; preference for 5' monophosphorylated substrate ''in vitro'', endonucleolytic cleavageNucleoside 2',3'-cyclic phosphate + H<sub>2</sub>O = nucleoside 3'-phosphate (according to Swiss-Prot) required for the processing of the ''[[gapA]]'' operon mRNA [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed], cleavage activity appears sensitive to downstream secondary structure. |
| − | * '''Protein family:''' 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot) 2',3' cyclic nucleotide phosphodiesterase family | + | * '''Protein family:''' Member of the HD superfamily of metal-dependent phosphohydrolases; 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot) 2',3' cyclic nucleotide phosphodiesterase family |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 74: | Line 74: | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' requires Mg+2, which can be replaced by Zn+2 or Mn+2 ions |
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' appeares sensitive to downstream secondary structure |
* '''Interactions:''' [[Rny]]-[[PfkA]], [[Rny]]-[[Eno]], [[Rny]]-[[PnpA]], [[Rny]]-[[RnjA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed] | * '''Interactions:''' [[Rny]]-[[PfkA]], [[Rny]]-[[Eno]], [[Rny]]-[[PnpA]], [[Rny]]-[[RnjA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed] | ||
| Line 109: | Line 109: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' essential!!!!, 4043 (''rny'' under p-spac control, ''cat''), GP193 (''rny'' under p-xyl control, ''cat''), both available in [[Stülke]] lab | + | * '''Mutant:''' essential!!!!, 4043 (''rny'' under p-spac control, ''cat''), GP193 (''rny'' under p-xyl control, ''cat''), both available in [[Stülke]] lab; SSB447 (rny under P-spac control, "erm") available in [[Putzer]] lab. |
* '''Expression vector:''' | * '''Expression vector:''' | ||
** N-terminal Strep-tag, expression in ''E. coli'', in [[pGP172]]: pGP441, available in [[Stülke]] lab | ** N-terminal Strep-tag, expression in ''E. coli'', in [[pGP172]]: pGP441, available in [[Stülke]] lab | ||
** N-terminal Strep-tag, for [[SPINE]], expression in ''B. subtilis'', in [[pGP380]]: pGP775 , available in [[Stülke]] lab | ** N-terminal Strep-tag, for [[SPINE]], expression in ''B. subtilis'', in [[pGP380]]: pGP775 , available in [[Stülke]] lab | ||
| + | **Expression of RNase Y missing the N-terminal transmembrane domain (25aa) as an intein fusion in E. coli (no tag left in the purified protein) available in the [[Putzer]] lab | ||
** wild type ''rny'', expression in ''B. subtilis'', in [[pBQ200]]: pGP1201, available in [[Stülke]] lab | ** wild type ''rny'', expression in ''B. subtilis'', in [[pBQ200]]: pGP1201, available in [[Stülke]] lab | ||
** there is also a series of domain constructs present in [[pBQ200]], all available in [[Stülke]] lab | ** there is also a series of domain constructs present in [[pBQ200]], all available in [[Stülke]] lab | ||
Revision as of 14:20, 5 October 2009
- Description: RNase Y, 5' end sensitive endoribonuclease, involved in the degradation/processing of mRNA
| Gene name | rny |
| Synonyms | ymdA |
| Essential | yes |
| Product | RNase Y |
| Function | Initiates S-box riboswitch RNA turnover, required for the processing of the gapA operon mRNA, depletion of RNase Y increases bulk mRNA stability. |
| Regulatory function of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 58,7 kDa, 5.39 |
| Gene length, protein length | 1560 bp, 520 amino acids |
| Immediate neighbours | pbpX, ymdB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 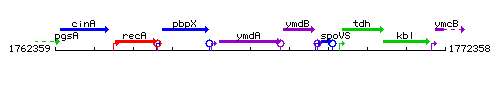
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU16960
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: RNase Y cleaves S-box riboswitch RNAs in vivo and in vitro; preference for 5' monophosphorylated substrate in vitro, endonucleolytic cleavageNucleoside 2',3'-cyclic phosphate + H2O = nucleoside 3'-phosphate (according to Swiss-Prot) required for the processing of the gapA operon mRNA PubMed, cleavage activity appears sensitive to downstream secondary structure.
- Protein family: Member of the HD superfamily of metal-dependent phosphohydrolases; 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot) 2',3' cyclic nucleotide phosphodiesterase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- transmembrane domain (4–24)
- KH domain (210–273)
- HD domain (336–429)
- Modification:
- Cofactor(s): requires Mg+2, which can be replaced by Zn+2 or Mn+2 ions
- Effectors of protein activity: appeares sensitive to downstream secondary structure
Database entries
- Structure:
- UniProt: O31774
- KEGG entry: [2]
- E.C. number: 3.1.4.16
Additional information
required for the processing of the gapA operon mRNA
Expression and regulation
- Sigma factor:
- Regulation: constitutive
- Regulatory mechanism:
- Additional information: there is a terminator between rny and ymdB, most transcripts terminate there
Biological materials
- Mutant: essential!!!!, 4043 (rny under p-spac control, cat), GP193 (rny under p-xyl control, cat), both available in Stülke lab; SSB447 (rny under P-spac control, "erm") available in Putzer lab.
- Expression vector:
- N-terminal Strep-tag, expression in E. coli, in pGP172: pGP441, available in Stülke lab
- N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380: pGP775 , available in Stülke lab
- Expression of RNase Y missing the N-terminal transmembrane domain (25aa) as an intein fusion in E. coli (no tag left in the purified protein) available in the Putzer lab
- wild type rny, expression in B. subtilis, in pBQ200: pGP1201, available in Stülke lab
- there is also a series of domain constructs present in pBQ200, all available in Stülke lab
- GFP fusion: B. subtilis 3569 (amyE:: (p-xyl rny-gfpmut1-spc)), available in Errington lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
Labs working on this gene/protein
Harald Putzer, IBPC Paris, France Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Karen Shahbabian, Ailar Jamalli, Léna Zig, Harald Putzer
RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis.
EMBO J: 2009, 28(22);3523-33
[PubMed:19779461]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)