Difference between revisions of "RnjA"
(→Biological materials) |
|||
| Line 123: | Line 123: | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| + | ** for chromosomal expression of RNase J1-Strep (spc): GP1034, available in [[Jörg Stülke]]'s lab | ||
| + | ** for chromosomal expression of RNase J1-Strep (cat): GP1042, available in [[Jörg Stülke]]'s lab | ||
* '''lacZ fusion:''' pGP418 (in [[pAC7]]), available in [[Stülke]] lab | * '''lacZ fusion:''' pGP418 (in [[pAC7]]), available in [[Stülke]] lab | ||
Revision as of 17:50, 18 April 2011
- Description: RNase J1
| Gene name | rnjA |
| Synonyms | ykqC |
| Essential | yes PubMed |
| Product | RNase J1 |
| Function | RNA processing |
| Metabolic function and regulation of this protein in SubtiPathways: Ile, Leu, Val, Coenzyme A | |
| MW, pI | 61 kDa, 5.902 |
| Gene length, protein length | 1665 bp, 555 aa |
| Immediate neighbours | adeC, ykzG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 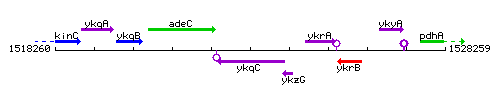
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14530
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: endonuclease and 5'-3' exonuclease
- Protein family: RNase J subfamily (according to Swiss-Prot)
- Paralogous protein(s): RnjB
RNAs affected by rnjA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: part of the RNA degradosome
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 3BK1 (RNase J from Thermus thermophilus) 3BK2 (RNase J from Thermus thermophilus, complex with UMP)
- UniProt: Q45493
- KEGG entry: [2]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- required for thrS RNA processing, involved in maturation of the 5’-end of the16S rRNA
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: GP41 (rnjA under control of p(xyl)), available in Stülke lab; SSB342 (rnjA under pspac), cat, available in Harald Putzer lab
- Expression vector:
- for chromosomal expression of RNase J1-Strep (spc): GP1034, available in Jörg Stülke's lab
- for chromosomal expression of RNase J1-Strep (cat): GP1042, available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Harald Putzer, IBPC Paris, France Homepage
David Bechhofer, Mount Sinai School, New York, USA Homepage
Ciaran Condon, IBPC, Paris, France Homepage
Your additional remarks
References
Reviews
Ciarán Condon, David H Bechhofer
Regulated RNA stability in the Gram positives.
Curr Opin Microbiol: 2011, 14(2);148-54
[PubMed:21334965]
[WorldCat.org]
[DOI]
(I p)
Ciarán Condon
What is the role of RNase J in mRNA turnover?
RNA Biol: 2010, 7(3);316-21
[PubMed:20458164]
[WorldCat.org]
[DOI]
(I p)
Original publications
Martin Lehnik-Habrink, Henrike Pförtner, Leonie Rempeters, Nico Pietack, Christina Herzberg, Jörg Stülke
The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex.
Mol Microbiol: 2010, 77(4);958-71
[PubMed:20572937]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, David H Bechhofer
Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y.
J Bacteriol: 2010, 192(13);3279-86
[PubMed:20418391]
[WorldCat.org]
[DOI]
(I p)
Nathalie Mathy, Agnès Hébert, Peggy Mervelet, Lionel Bénard, Audrey Dorléans, Inés Li de la Sierra-Gallay, Philippe Noirot, Harald Putzer, Ciarán Condon
Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour.
Mol Microbiol: 2010, 75(2);489-98
[PubMed:20025672]
[WorldCat.org]
[DOI]
(I p)
Yulia Redko, Ciarán Condon
Maturation of 23S rRNA in Bacillus subtilis in the absence of Mini-III.
J Bacteriol: 2010, 192(1);356-9
[PubMed:19880604]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, Josh S Sharp, David H Bechhofer
Bacillus subtilis RNase J1 endonuclease and 5' exonuclease activities in the turnover of DeltaermC mRNA.
RNA: 2009, 15(12);2331-9
[PubMed:19850915]
[WorldCat.org]
[DOI]
(I p)
Gintaras Deikus, David H Bechhofer
Bacillus subtilis trp Leader RNA: RNase J1 endonuclease cleavage specificity and PNPase processing.
J Biol Chem: 2009, 284(39);26394-401
[PubMed:19638340]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, David H Bechhofer
Processing and stability of inducibly expressed rpsO mRNA derivatives in Bacillus subtilis.
J Bacteriol: 2009, 191(18);5680-9
[PubMed:19633085]
[WorldCat.org]
[DOI]
(I p)
Ming Fang, Wencke-Maria Zeisberg, Ciaran Condon, Vasily Ogryzko, Antoine Danchin, Undine Mechold
Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis.
Nucleic Acids Res: 2009, 37(15);5114-25
[PubMed:19553197]
[WorldCat.org]
[DOI]
(I p)
Roula Daou-Chabo, Ciarán Condon
RNase J1 endonuclease activity as a probe of RNA secondary structure.
RNA: 2009, 15(7);1417-25
[PubMed:19458035]
[WorldCat.org]
[DOI]
(I p)
Roula Daou-Chabo, Nathalie Mathy, Lionel Bénard, Ciarán Condon
Ribosomes initiating translation of the hbs mRNA protect it from 5'-to-3' exoribonucleolytic degradation by RNase J1.
Mol Microbiol: 2009, 71(6);1538-50
[PubMed:19210617]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Léna Zig, Julia Kretschmer, Georg Homuth, Harald Putzer
mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale.
Mol Microbiol: 2008, 70(1);183-96
[PubMed:18713320]
[WorldCat.org]
[DOI]
(I p)
Gintaras Deikus, Ciarán Condon, David H Bechhofer
Role of Bacillus subtilis RNase J1 endonuclease and 5'-exonuclease activities in trp leader RNA turnover.
J Biol Chem: 2008, 283(25);17158-67
[PubMed:18445592]
[WorldCat.org]
[DOI]
(P p)
Inés Li de la Sierra-Gallay, Léna Zig, Ailar Jamalli, Harald Putzer
Structural insights into the dual activity of RNase J.
Nat Struct Mol Biol: 2008, 15(2);206-12
[PubMed:18204464]
[WorldCat.org]
[DOI]
(I p)
Jennifer A Collins, Irnov Irnov, Stephanie Baker, Wade C Winkler
Mechanism of mRNA destabilization by the glmS ribozyme.
Genes Dev: 2007, 21(24);3356-68
[PubMed:18079181]
[WorldCat.org]
[DOI]
(P p)
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, Joshua B Blaustein, David H Bechhofer
Processing of Bacillus subtilis small cytoplasmic RNA: evidence for an additional endonuclease cleavage site.
Nucleic Acids Res: 2007, 35(13);4464-73
[PubMed:17576666]
[WorldCat.org]
[DOI]
(I p)
Nathalie Mathy, Lionel Bénard, Olivier Pellegrini, Roula Daou, Tingyi Wen, Ciarán Condon
5'-to-3' exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5' stability of mRNA.
Cell: 2007, 129(4);681-92
[PubMed:17512403]
[WorldCat.org]
[DOI]
(P p)
Robert A Britton, Tingyi Wen, Laura Schaefer, Olivier Pellegrini, William C Uicker, Nathalie Mathy, Crystal Tobin, Roula Daou, Jacek Szyk, Ciarán Condon
Maturation of the 5' end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1.
Mol Microbiol: 2007, 63(1);127-38
[PubMed:17229210]
[WorldCat.org]
[DOI]
(P p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)
Sergine Even, Olivier Pellegrini, Lena Zig, Valerie Labas, Joelle Vinh, Dominique Bréchemmier-Baey, Harald Putzer
Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E.
Nucleic Acids Res: 2005, 33(7);2141-52
[PubMed:15831787]
[WorldCat.org]
[DOI]
(I e)