RibE
- Description: riboflavin synthase (alpha subunit)
| Gene name | ribE |
| Synonyms | ribB |
| Essential | no |
| Product | riboflavin synthase (alpha subunit) |
| Function | riboflavin biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Riboflavin / FAD | |
| MW, pI | 23 kDa, 5.845 |
| Gene length, protein length | 645 bp, 215 aa |
| Immediate neighbours | ribA, ribD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 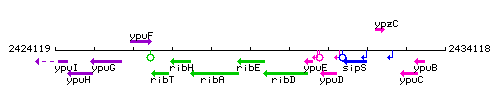
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23270
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Categories containing this gene/protein
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 6,7-dimethyl-8-(1-D-ribityl)lumazine = riboflavin + 4-(1-D-ribitylamino)-5-amino-2,6-dihydroxypyrimidine (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1I8D (from E. coli, 36% identity, 59% similarity) PubMed, , 1RVV (the RibE)3-(RibH)60 lumazine synthase/riboflavin synthase complex) PubMed
- UniProt: P16440
- KEGG entry: [3]
- E.C. number: 2.5.1.9
Additional information
Expression and regulation
- Regulatory mechanism: FMN-box: riboswitch, mediates termination/ antitermination control of the operon, in the absence of FMN: antitermination, in the presence of FMN: termination PubMed
- Additional information:
Biological materials
- Mutant: GP203 (erm), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
J Kenneth Wickiser, Wade C Winkler, Ronald R Breaker, Donald M Crothers
The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch.
Mol Cell: 2005, 18(1);49-60
[PubMed:15808508]
[WorldCat.org]
[DOI]
(P p)
Wade C Winkler, Smadar Cohen-Chalamish, Ronald R Breaker
An mRNA structure that controls gene expression by binding FMN.
Proc Natl Acad Sci U S A: 2002, 99(25);15908-13
[PubMed:12456892]
[WorldCat.org]
[DOI]
(P p)
D I Liao, Z Wawrzak, J C Calabrese, P V Viitanen, D B Jordan
Crystal structure of riboflavin synthase.
Structure: 2001, 9(5);399-408
[PubMed:11377200]
[WorldCat.org]
[DOI]
(P p)
K Ritsert, R Huber, D Turk, R Ladenstein, K Schmidt-Bäse, A Bacher
Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J Mol Biol: 1995, 253(1);151-67
[PubMed:7473709]
[WorldCat.org]
[DOI]
(P p)
V N Mironov, A S Kraev, M L Chikindas, B K Chernov, A I Stepanov, K G Skryabin
Functional organization of the riboflavin biosynthesis operon from Bacillus subtilis SHgw.
Mol Gen Genet: 1994, 242(2);201-8
[PubMed:8159171]
[WorldCat.org]
[DOI]
(P p)
V Azevedo, A Sorokin, S D Ehrlich, P Serror
The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci.
Mol Microbiol: 1993, 10(2);397-405
[PubMed:7934830]
[WorldCat.org]
[DOI]
(P p)