Difference between revisions of "RhiN"
| Line 51: | Line 51: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU07000&redirect=T BSU07000] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 86: | Line 87: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU07000&redirect=T BSU07000] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 13:09, 2 April 2014
- Description: galacturonyl hydrolase, catalyses intracellular degradation of disaccharides generated by YesX
| Gene name | yesR |
| Synonyms | |
| Essential | no |
| Product | galacturonyl hydrolase, catalyses intracellular degradation of disaccharides generated by YesX |
| Function | intracellular degradation of disaccharides generated by YesX |
| Gene expression levels in SubtiExpress: yesR | |
| MW, pI | 38 kDa, 4.757 |
| Gene length, protein length | 1032 bp, 344 aa |
| Immediate neighbours | yesQ, yesS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 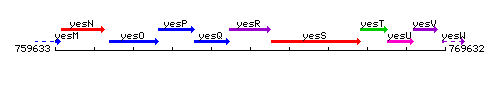
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed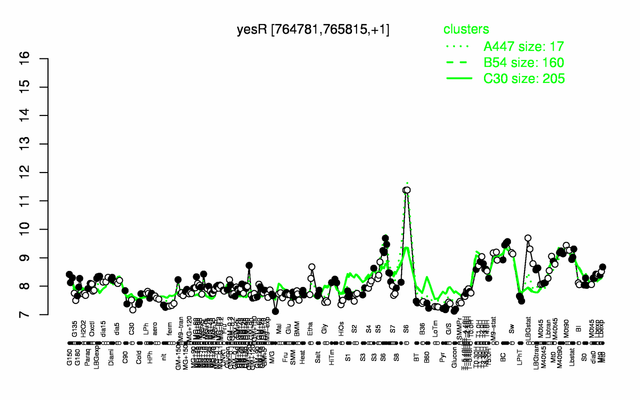
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU07000
Phenotypes of a mutant
Database entries
- BsubCyc: BSU07000
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: glycosyl hydrolase 105 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU07000
- Structure:
- UniProt: O31521
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- induced by pectin PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irina A Rodionova, Xiaoqing Li, Vera Thiel, Sergey Stolyar, Krista Stanton, James K Fredrickson, Donald A Bryant, Andrei L Osterman, Aaron A Best, Dmitry A Rodionov
Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria.
Front Microbiol: 2013, 4;407
[PubMed:24391637]
[WorldCat.org]
[DOI]
(P e)
Akihito Ochiai, Takafumi Itoh, Akiko Kawamata, Wataru Hashimoto, Kousaku Murata
Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization.
Appl Environ Microbiol: 2007, 73(12);3803-13
[PubMed:17449691]
[WorldCat.org]
[DOI]
(P p)
Takafumi Itoh, Akihito Ochiai, Bunzo Mikami, Wataru Hashimoto, Kousaku Murata
A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide.
J Mol Biol: 2006, 360(3);573-85
[PubMed:16781735]
[WorldCat.org]
[DOI]
(P p)