Difference between revisions of "RhiL"
(→Extended information on the protein) |
|||
| Line 90: | Line 90: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' ([[YesP]]-[[YesQ]])-[[YesO]] {{PubMed|10092453}} | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** ([[YesP]]-[[YesQ]])-[[YesO]] {{PubMed|10092453}} | ||
| − | * '''Localization:''' associated to the membrane (via [[YesP]]-[[YesQ]]) {{PubMed|10092453}} | + | * '''[[Localization]]:''' |
| + | ** associated to the membrane (via [[YesP]]-[[YesQ]]) {{PubMed|10092453}} | ||
=== Database entries === | === Database entries === | ||
Revision as of 18:31, 29 July 2012
- Description: similar to ABC transporter (sugar-binding protein)
| Gene name | yesO |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Interactions involving this protein in SubtInteract: YesO | |
| MW, pI | 46 kDa, 4.992 |
| Gene length, protein length | 1236 bp, 412 aa |
| Immediate neighbours | yesN, yesP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 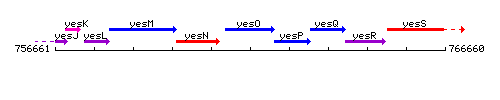
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed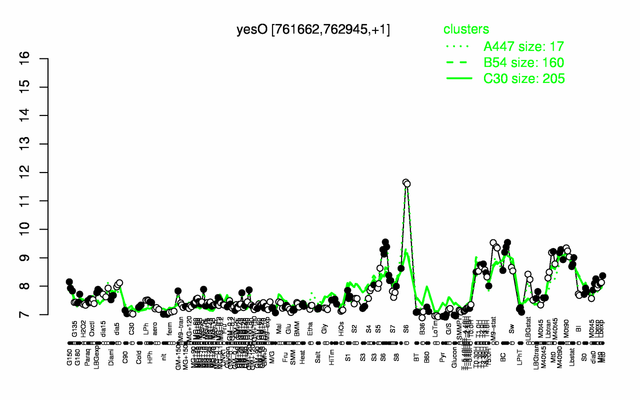
| |
Contents
Categories containing this gene/protein
ABC transporters, utilization of specific carbon sources, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06970
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial solute-binding protein 1 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31518
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- induced by pectin PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Akihito Ochiai, Takafumi Itoh, Akiko Kawamata, Wataru Hashimoto, Kousaku Murata
Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization.
Appl Environ Microbiol: 2007, 73(12);3803-13
[PubMed:17449691]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)