Difference between revisions of "RhgR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcriptional activator (AraC family) of the rhamnogalacturonan operon <br/><br/> | + | * '''Description:''' transcriptional activator ([[transcription factors of the AraC family|AraC family]]) of the rhamnogalacturonan operon <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''rhgR'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''yesS '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 13: | Line 13: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of pectin utilization | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of pectin utilization | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU07010 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU07010 rhgR] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein= | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=RhgR RhgR] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 87 kDa, 7.77 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 87 kDa, 7.77 | ||
| Line 21: | Line 21: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2283 bp, 761 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2283 bp, 761 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rhiN]]'', ''[[yesT]]'' |
|- | |- | ||
|style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU07010 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU07010 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU07010 DNA_with_flanks] | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU07010 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU07010 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU07010 DNA_with_flanks] | ||
| Line 44: | Line 44: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[ | + | {{SubtiWiki regulon|[[RhgR regulon]]}} |
| − | =The [[ | + | =The [[RhgR regulon]]:= |
| − | * ''[[ | + | * ''[[rhgR]]-[[yesT]]-[[yesU]]-[[yesV]]-[[yesW]]-[[yesX]]-[[yesY]]-[[yesZ]]'' |
=The gene= | =The gene= | ||
| Line 88: | Line 88: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** [[ | + | ** [[RhgR]] is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of [[SubtInteract|interactions]] {{PubMed|21630458}} |
| − | ** [[PtsH|HPr(His)-P]]-[[ | + | ** [[PtsH|HPr(His)-P]]-[[RhgR]] {{PubMed|19651770}} |
* '''[[Localization]]:''' cell membrane (according to Swiss-Prot) | * '''[[Localization]]:''' cell membrane (according to Swiss-Prot) | ||
| Line 108: | Line 108: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[rhgR]]-[[yesT]]-[[yesU]]-[[yesV]]-[[yesW]]-[[yesX]]-[[yesY]]-[[yesZ]]'' {{PubMed|19651770}} |
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yesS_765838_768123_1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yesS_765838_768123_1 rhgR] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| − | * '''Regulation:''' induced by pectin ([[ | + | * '''Regulation:''' induced by pectin ([[RhgR]]) {{PubMed|19651770}} |
| − | * '''Regulatory mechanism:''' [[ | + | * '''Regulatory mechanism:''' [[RhgR]]: transcriptional activation {{PubMed|19651770}} |
* '''Additional information:''' | * '''Additional information:''' | ||
Revision as of 15:54, 7 June 2014
- Description: transcriptional activator (AraC family) of the rhamnogalacturonan operon
| Gene name | rhgR |
| Synonyms | yesS |
| Essential | no |
| Product | transcriptional activator of the rhamnogalacturonan operon |
| Function | regulation of pectin utilization |
| Gene expression levels in SubtiExpress: rhgR | |
| Interactions involving this protein in SubtInteract: RhgR | |
| MW, pI | 87 kDa, 7.77 |
| Gene length, protein length | 2283 bp, 761 aa |
| Immediate neighbours | rhiN, yesT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 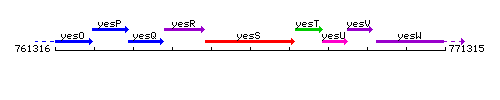
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed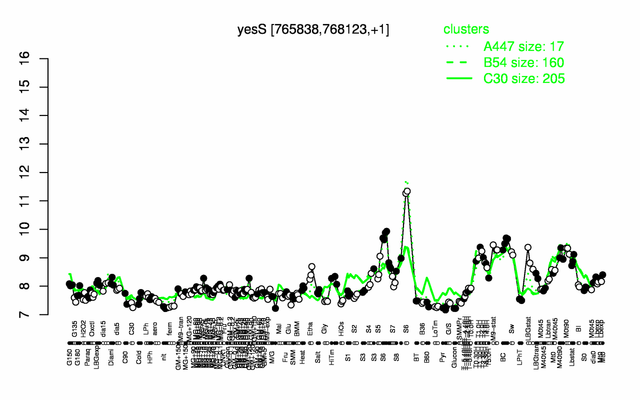
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control, membrane proteins
This gene is a member of the following regulons
The RhgR regulon:
The gene
Basic information
- Locus tag: BSU07010
Phenotypes of a mutant
Database entries
- BsubCyc: BSU07010
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity: interaction with HPr(His)-P (occurs in the absence of preferred carbon sources) stimulates YesS activity PubMed
- Interactions:
- RhgR is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of interactions PubMed
- HPr(His)-P-RhgR PubMed
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU07010
- Structure:
- UniProt: O31522
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irina A Rodionova, Xiaoqing Li, Vera Thiel, Sergey Stolyar, Krista Stanton, James K Fredrickson, Donald A Bryant, Andrei L Osterman, Aaron A Best, Dmitry A Rodionov
Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria.
Front Microbiol: 2013, 4;407
[PubMed:24391637]
[WorldCat.org]
[DOI]
(P e)
Elodie Marchadier, Rut Carballido-López, Sophie Brinster, Céline Fabret, Peggy Mervelet, Philippe Bessières, Marie-Françoise Noirot-Gros, Vincent Fromion, Philippe Noirot
An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach.
Proteomics: 2011, 11(15);2981-91
[PubMed:21630458]
[WorldCat.org]
[DOI]
(I p)
Sandrine Poncet, Maryline Soret, Peggy Mervelet, Josef Deutscher, Philippe Noirot
Transcriptional activator YesS is stimulated by histidine-phosphorylated HPr of the Bacillus subtilis phosphotransferase system.
J Biol Chem: 2009, 284(41);28188-28197
[PubMed:19651770]
[WorldCat.org]
[DOI]
(I p)
Akihito Ochiai, Takafumi Itoh, Akiko Kawamata, Wataru Hashimoto, Kousaku Murata
Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization.
Appl Environ Microbiol: 2007, 73(12);3803-13
[PubMed:17449691]
[WorldCat.org]
[DOI]
(P p)