Difference between revisions of "Rex"
Raphael2215 (talk | contribs) |
|||
| Line 36: | Line 36: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| Line 107: | Line 103: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[ydiG]]-[[rex]]-[[tatAY]]-[[tatCY]]'' {{PubMed|22383849}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rex_647091_647738_1 rex] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rex_647091_647738_1 rex] {{PubMed|22383849}} | ||
| Line 142: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>16207915,18485070,15231791,17015645,20525796, 21402078 </pubmed> | + | <pubmed>16207915,18485070,15231791,17015645,20525796, 21402078 22383849</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:08, 28 August 2012
- Description: transcriptional repressor of anaerobically expressed genes involved in anaerobic respiration and fermentation
| Gene name | rex |
| Synonyms | ydiH |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of fermentation and anaerobic respiration |
| Gene expression levels in SubtiExpress: rex | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 23 kDa, 6.772 |
| Gene length, protein length | 645 bp, 215 aa |
| Immediate neighbours | ydiG, tatAY |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 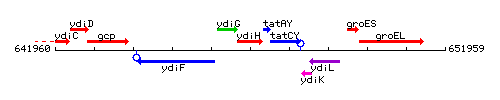
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed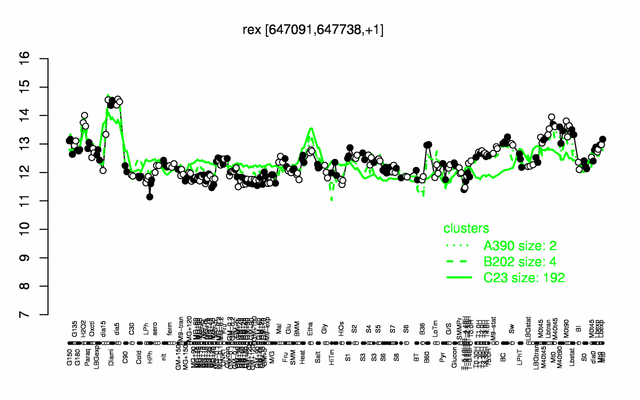
| |
Contents
Categories containing this gene/protein
regulators of electron transport, carbon core metabolism, transcription factors and their control
This gene is a member of the following regulons
The Rex regulon
The gene
Basic information
- Locus tag: BSU05970
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: transcriptional regulatory rex family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: NADH (indication of low oxygen concentration) releases Rex from DNA PubMed, Rex has a very highh affinity for NADH
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: O05521
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant: LUW292 (erm), GP834 (erm), both available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Claes von Wachenfeldt, Lund University, Sweden Homepage
Your additional remarks
References
Pierre Nicolas, Ulrike Mäder, Etienne Dervyn, Tatiana Rochat, Aurélie Leduc, Nathalie Pigeonneau, Elena Bidnenko, Elodie Marchadier, Mark Hoebeke, Stéphane Aymerich, Dörte Becher, Paola Bisicchia, Eric Botella, Olivier Delumeau, Geoff Doherty, Emma L Denham, Mark J Fogg, Vincent Fromion, Anne Goelzer, Annette Hansen, Elisabeth Härtig, Colin R Harwood, Georg Homuth, Hanne Jarmer, Matthieu Jules, Edda Klipp, Ludovic Le Chat, François Lecointe, Peter Lewis, Wolfram Liebermeister, Anika March, Ruben A T Mars, Priyanka Nannapaneni, David Noone, Susanne Pohl, Bernd Rinn, Frank Rügheimer, Praveen K Sappa, Franck Samson, Marc Schaffer, Benno Schwikowski, Leif Steil, Jörg Stülke, Thomas Wiegert, Kevin M Devine, Anthony J Wilkinson, Jan Maarten van Dijl, Michael Hecker, Uwe Völker, Philippe Bessières, Philippe Noirot
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
Science: 2012, 335(6072);1103-6
[PubMed:22383849]
[WorldCat.org]
[DOI]
(I p)
Ellen Wang, Teemu P Ikonen, Matti Knaapila, Dmitri Svergun, Derek T Logan, Claes von Wachenfeldt
Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD+ binding in solution.
J Mol Biol: 2011, 408(4);670-83
[PubMed:21402078]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Ellen Wang, Mikael C Bauer, Annika Rogstam, Sara Linse, Derek T Logan, Claes von Wachenfeldt
Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex.
Mol Microbiol: 2008, 69(2);466-78
[PubMed:18485070]
[WorldCat.org]
[DOI]
(I p)
Smita Gyan, Yoshihiko Shiohira, Ichiro Sato, Michio Takeuchi, Tsutomu Sato
Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis.
J Bacteriol: 2006, 188(20);7062-71
[PubMed:17015645]
[WorldCat.org]
[DOI]
(P p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 10);3323-3335
[PubMed:16207915]
[WorldCat.org]
[DOI]
(P p)
Matthew Schau, Yinghua Chen, F Marion Hulett
Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon.
J Bacteriol: 2004, 186(14);4585-95
[PubMed:15231791]
[WorldCat.org]
[DOI]
(P p)