Difference between revisions of "Rex"
| Line 1: | Line 1: | ||
| − | + | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of fermentation and anaerobic respiration | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of fermentation and anaerobic respiration | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU05970 rex] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
Revision as of 15:33, 6 August 2012
| Gene name | rex |
| Synonyms | ydiH |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of fermentation and anaerobic respiration |
| Gene expression levels in SubtiExpress: rex | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 23 kDa, 6.772 |
| Gene length, protein length | 645 bp, 215 aa |
| Immediate neighbours | ydiG, tatAY |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 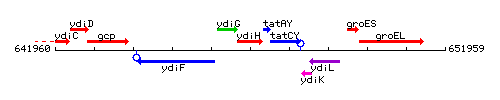
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed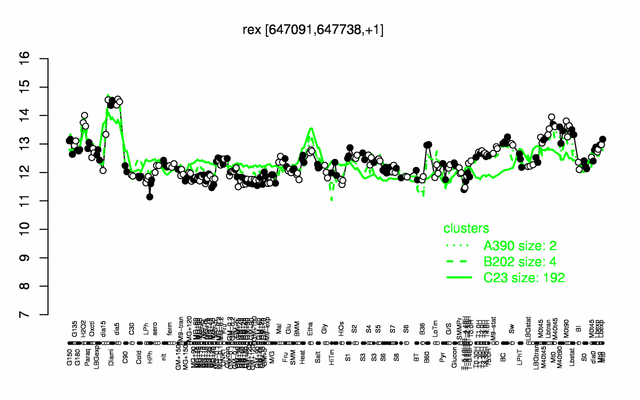
| |
Contents
Categories containing this gene/protein
regulators of electron transport, carbon core metabolism, transcription factors and their control
This gene is a member of the following regulons
The Rex regulon
The gene
Basic information
- Locus tag: BSU05970
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: transcriptional regulatory rex family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: NADH (indication of low oxygen concentration) releases Rex from DNA PubMed, Rex has a very highh affinity for NADH
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: O05521
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant: LUW292 (erm), GP834 (erm), both available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Claes von Wachenfeldt, Lund University, Sweden Homepage
Your additional remarks
References
Ellen Wang, Teemu P Ikonen, Matti Knaapila, Dmitri Svergun, Derek T Logan, Claes von Wachenfeldt
Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD+ binding in solution.
J Mol Biol: 2011, 408(4);670-83
[PubMed:21402078]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Ellen Wang, Mikael C Bauer, Annika Rogstam, Sara Linse, Derek T Logan, Claes von Wachenfeldt
Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex.
Mol Microbiol: 2008, 69(2);466-78
[PubMed:18485070]
[WorldCat.org]
[DOI]
(I p)
Smita Gyan, Yoshihiko Shiohira, Ichiro Sato, Michio Takeuchi, Tsutomu Sato
Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis.
J Bacteriol: 2006, 188(20);7062-71
[PubMed:17015645]
[WorldCat.org]
[DOI]
(P p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 10);3323-3335
[PubMed:16207915]
[WorldCat.org]
[DOI]
(P p)
Matthew Schau, Yinghua Chen, F Marion Hulett
Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon.
J Bacteriol: 2004, 186(14);4585-95
[PubMed:15231791]
[WorldCat.org]
[DOI]
(P p)