Difference between revisions of "RemB"
| Line 44: | Line 44: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | ** smooth colonies on MsGG medium, no [[biofilm formation]] {{PubMed|22113911}} | ||
=== Database entries === | === Database entries === | ||
| Line 128: | Line 129: | ||
=References= | =References= | ||
| − | <pubmed>,19363116, 19363116, 2987848 </pubmed> | + | <pubmed>,19363116, 19363116, 2987848 22113911</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:44, 25 November 2011
- Description: involved in the activation of biofilm matrix biosynthetic operons, acts in parallel to SinR, AbrB, and DegU
| Gene name | remB |
| Synonyms | yaaB |
| Essential | no |
| Product | regulator of the extracellular matrix |
| Function | regulation of biofilm formation |
| MW, pI | 5 kDa, 8.796 |
| Gene length, protein length | 156 bp, 52 aa |
| Immediate neighbours | recF, gyrB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 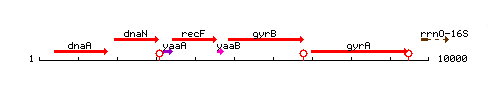
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00050
Phenotypes of a mutant
- smooth colonies on MsGG medium, no biofilm formation PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P37525
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Daniel Kearns, Indiana University, Bloomington, USA, homepage
Your additional remarks
References
Eric R Pozsgai, Kris M Blair, Daniel B Kearns
Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis.
Appl Environ Microbiol: 2012, 78(3);778-85
[PubMed:22113911]
[WorldCat.org]
[DOI]
(I p)
Jared T Winkelman, Kris M Blair, Daniel B Kearns
RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis.
J Bacteriol: 2009, 191(12);3981-91
[PubMed:19363116]
[WorldCat.org]
[DOI]
(I p)
N Ogasawara, S Moriya, H Yoshikawa
Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.
Nucleic Acids Res: 1985, 13(7);2267-79
[PubMed:2987848]
[WorldCat.org]
[DOI]
(P p)