Difference between revisions of "RecU"
(→Biological materials) |
|||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || DNA repair, homologous recombination and chromosome segregation | |style="background:#ABCDEF;" align="center"|'''Function''' || DNA repair, homologous recombination and chromosome segregation | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/RecU RecU] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 23 kDa, 9.511 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 23 kDa, 9.511 | ||
Revision as of 19:29, 31 July 2011
- Description: DNA repair, homologous recombination and chromosome segregation; recognizes, distorts, and cleaves four-stranded recombination intermediates and modulates RecA activities
| Gene name | recU |
| Synonyms | recG, prfA, yppB, jopB |
| Essential | no |
| Product | Holliday junction resolvase |
| Function | DNA repair, homologous recombination and chromosome segregation |
| Interactions involving this protein in SubtInteract: RecU | |
| MW, pI | 23 kDa, 9.511 |
| Gene length, protein length | 618 bp, 206 aa |
| Immediate neighbours | yppC, ponA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 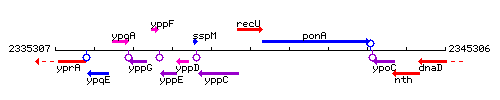
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, cell envelope stress proteins (controlled by SigM, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1ZP7
- UniProt: P39792
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP891 (recU::cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Cristina Cañas, Begoña Carrasco, Esther García-Tirado, John B Rafferty, Juan C Alonso, Silvia Ayora
The stalk region of the RecU resolvase is essential for Holliday junction recognition and distortion.
J Mol Biol: 2011, 410(1);39-49
[PubMed:21600217]
[WorldCat.org]
[DOI]
(I p)
Dawit Kidane, Begoña Carrasco, Candela Manfredi, Katharina Rothmaier, Silvia Ayora, Serkalem Tadesse, Juan C Alonso, Peter L Graumann
Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells.
PLoS Genet: 2009, 5(9);e1000630
[PubMed:19730681]
[WorldCat.org]
[DOI]
(I p)
Begoña Carrasco, Cristina Cañas, Gary J Sharples, Juan C Alonso, Silvia Ayora
The N-terminal region of the RecU holliday junction resolvase is essential for homologous recombination.
J Mol Biol: 2009, 390(1);1-9
[PubMed:19422832]
[WorldCat.org]
[DOI]
(I p)
Cristina Cañas, Begoña Carrasco, Silvia Ayora, Juan C Alonso
The RecU Holliday junction resolvase acts at early stages of homologous recombination.
Nucleic Acids Res: 2008, 36(16);5242-9
[PubMed:18684995]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Natalie McGregor, Sylvia Ayora, Svetlana Sedelnikova, Begona Carrasco, Juan C Alonso, Paul Thaw, John Rafferty
The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage.
Structure: 2005, 13(9);1341-51
[PubMed:16154091]
[WorldCat.org]
[DOI]
(P p)
Begoña Carrasco, Silvia Ayora, Rudi Lurz, Juan C Alonso
Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities.
Nucleic Acids Res: 2005, 33(12);3942-52
[PubMed:16024744]
[WorldCat.org]
[DOI]
(I e)
Humberto Sanchez, Dawit Kidane, Patricia Reed, Fiona A Curtis, M Castillo Cozar, Peter L Graumann, Gary J Sharples, Juan C Alonso
The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis.
Genetics: 2005, 171(3);873-83
[PubMed:16020779]
[WorldCat.org]
[DOI]
(P p)
Begoña Carrasco, M Castillo Cozar, Rudi Lurz, Juan C Alonso, Silvia Ayora
Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation.
J Bacteriol: 2004, 186(17);5557-66
[PubMed:15317759]
[WorldCat.org]
[DOI]
(P p)
B Carrasco, S Fernández, K Asai, N Ogasawara, J C Alonso
Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis.
Mol Genet Genomics: 2002, 266(5);899-906
[PubMed:11810266]
[WorldCat.org]
[DOI]
(P p)
S Fernández, A Sorokin, J C Alonso
Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination.
J Bacteriol: 1998, 180(13);3405-9
[PubMed:9642195]
[WorldCat.org]
[DOI]
(P p)
D L Popham, P Setlow
Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor.
J Bacteriol: 1995, 177(2);326-35
[PubMed:7814321]
[WorldCat.org]
[DOI]
(P p)