Difference between revisions of "RecJ"
| Line 4: | Line 4: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''recJ'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''yrvE'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || DNA repair | |style="background:#ABCDEF;" align="center"|'''Function''' || DNA repair | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU27620 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU27620 recJ] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=YrvE | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=YrvE RecJ] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=yrvE | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=yrvE recJ]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 87 kDa, 5.181 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 87 kDa, 5.181 | ||
| Line 91: | Line 91: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** [[SsbA]]-[[ | + | ** [[SsbA]]-[[RecJ]] {{PubMed|21170359}} |
* '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) | * '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) | ||
| Line 110: | Line 110: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[recJ]]-[[apt]]'' {{PubMed|15241682}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=recJ_2823419_2825779_-1 yrvE] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=recJ_2823419_2825779_-1 yrvE] {{PubMed|22383849}} | ||
| Line 127: | Line 127: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| − | ** GP895 (''recJ''::''kan''), available in [[Stülke]] lab | + | ** GP895 (''[[recJ]]''::''kan''), available in [[Jörg Stülke]]'s lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
Revision as of 11:06, 7 July 2015
- Description: single-strand DNA-specific exonuclease
| Gene name | recJ |
| Synonyms | yrvE |
| Essential | no |
| Product | single-strand DNA-specific exonuclease |
| Function | DNA repair |
| Gene expression levels in SubtiExpress: recJ | |
| Interactions involving this protein in SubtInteract: RecJ | |
| Metabolic function and regulation of this protein in SubtiPathways: recJ | |
| MW, pI | 87 kDa, 5.181 |
| Gene length, protein length | 2358 bp, 786 aa |
| Immediate neighbours | apt, yrvD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 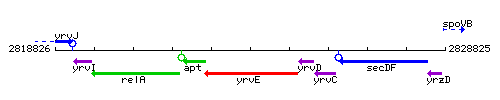
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
stringent response, Efp-dependent proteins
The gene
Basic information
- Locus tag: BSU27620
Phenotypes of a mutant
Database entries
- BsubCyc: BSU27620
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: recJ family (according to Swiss-Prot)
- Paralogous protein(s): YorK
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU27620
- Structure:
- UniProt: O32044
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
Biological materials
- Mutant:
- GP895 (recJ::kan), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Original publications
Hanjing Yang, Cameron Sikavi, Katherine Tran, Shauna M McGillivray, Victor Nizet, Madeline Yung, Aileen Chang, Jeffrey H Miller
Papillation in Bacillus anthracis colonies: a tool for finding new mutators.
Mol Microbiol: 2011, 79(5);1276-93
[PubMed:21205011]
[WorldCat.org]
[DOI]
(I p)
Audrey Costes, François Lecointe, Stephen McGovern, Sophie Quevillon-Cheruel, Patrice Polard
The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks.
PLoS Genet: 2010, 6(12);e1001238
[PubMed:21170359]
[WorldCat.org]
[DOI]
(I e)
M Nickel, G Homuth, C Böhnisch, U Mäder, T Schweder
Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability.
Mol Genet Genomics: 2004, 272(1);98-107
[PubMed:15241682]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
V A Sutera, E S Han, L A Rajman, S T Lovett
Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs.
J Bacteriol: 1999, 181(19);6098-102
[PubMed:10498723]
[WorldCat.org]
[DOI]
(P p)