RbsD
- Description: ribose ABC transporter (membrane protein)
| Gene name | rbsD |
| Synonyms | |
| Essential | no |
| Product | ribose ABC transporter (membrane protein) |
| Function | ribose uptake |
| Gene expression levels in SubtiExpress: rbsD | |
| Interactions involving this protein in SubtInteract: RbsD | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 14 kDa, 4.987 |
| Gene length, protein length | 393 bp, 131 aa |
| Immediate neighbours | rbsK, rbsA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 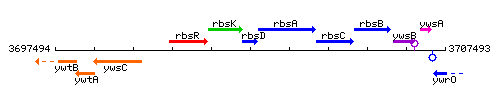
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed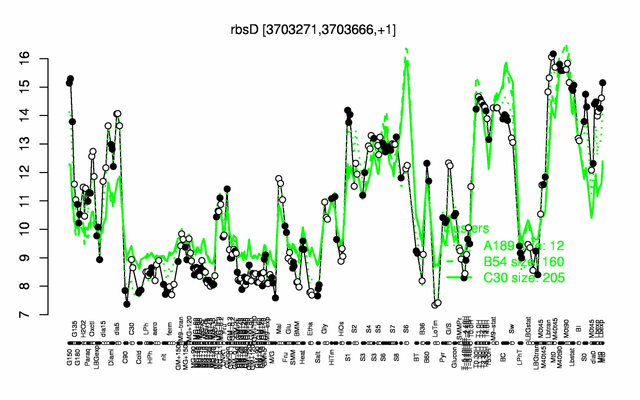
| |
Contents
Categories containing this gene/protein
ABC transporters, utilization of specific carbon sources, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35930
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: RbsD subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane PubMed
Database entries
- Structure: 1OGD (complex with D-ribose)
- UniProt: P36946
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Le Thi Tam, Christine Eymann, Dirk Albrecht, Rabea Sietmann, Frieder Schauer, Michael Hecker, Haike Antelmann
Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis.
Environ Microbiol: 2006, 8(8);1408-27
[PubMed:16872404]
[WorldCat.org]
[DOI]
(P p)
Min-Sung Kim, Joon Shin, Weontae Lee, Heung-Soo Lee, Byung-Ha Oh
Crystal structures of RbsD leading to the identification of cytoplasmic sugar-binding proteins with a novel folding architecture.
J Biol Chem: 2003, 278(30);28173-80
[PubMed:12738765]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon.
J Bacteriol: 1995, 177(23);6727-31
[PubMed:7592460]
[WorldCat.org]
[DOI]
(P p)
K Woodson, K M Devine
Analysis of a ribose transport operon from Bacillus subtilis.
Microbiology (Reading): 1994, 140 ( Pt 8);1829-38
[PubMed:7921236]
[WorldCat.org]
[DOI]
(P p)
M O'Reilly, K Woodson, B C Dowds, K M Devine
The citrulline biosynthetic operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilis are negatively regulated by Spo0A.
Mol Microbiol: 1994, 11(1);87-98
[PubMed:7511775]
[WorldCat.org]
[DOI]
(P p)