RacE
- Description: glutamate racemase
| Gene name | racE |
| Synonyms | glr |
| Essential | yes PubMed |
| Product | glutamate racemase |
| Function | peptidoglycan precursor biosynthesis |
| Gene expression levels in SubtiExpress: racE | |
| Metabolic function and regulation of this protein in SubtiPathways: Cell wall | |
| MW, pI | 29 kDa, 4.858 |
| Gene length, protein length | 816 bp, 272 aa |
| Immediate neighbours | gerM, ysmB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 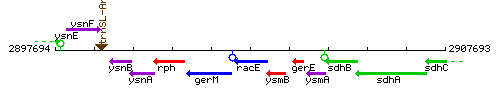
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28390
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate = D-glutamate (according to Swiss-Prot)
- Protein family: aspartate/glutamate racemases family (according to Swiss-Prot)
- Paralogous protein(s): yrpC
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1ZUW (with D-glutamate)
- UniProt: P94556
- KEGG entry: [2]
- E.C. number: 5.1.1.3
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
M Ashley Spies, Joseph G Reese, Dylan Dodd, Katherine L Pankow, Steven R Blanke, Jerome Baudry
Determinants of catalytic power and ligand binding in glutamate racemase.
J Am Chem Soc: 2009, 131(14);5274-84
[PubMed:19309142]
[WorldCat.org]
[DOI]
(I p)
Eduard Puig, Edgar Mixcoha, Mireia Garcia-Viloca, Angels González-Lafont, José M Lluch
How the substrate D-glutamate drives the catalytic action of Bacillus subtilis glutamate racemase.
J Am Chem Soc: 2009, 131(10);3509-21
[PubMed:19227983]
[WorldCat.org]
[DOI]
(I p)
Sergey N Ruzheinikov, Makie A Taal, Svetlana E Sedelnikova, Patrick J Baker, David W Rice
Substrate-induced conformational changes in Bacillus subtilis glutamate racemase and their implications for drug discovery.
Structure: 2005, 13(11);1707-13
[PubMed:16271894]
[WorldCat.org]
[DOI]
(P p)
Keitarou Kimura, Lam-Son Phan Tran, Yoshifumi Itoh
Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis.
Microbiology (Reading): 2004, 150(Pt 9);2911-2920
[PubMed:15347750]
[WorldCat.org]
[DOI]
(P p)