Difference between revisions of "QcrB"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 111: | Line 107: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=qcrB_2363913_2364587_-1 qcrB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=qcrB_2363913_2364587_-1 qcrB] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|7592464}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|7592464}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 144: | Line 140: | ||
=References= | =References= | ||
| − | + | <pubmed>8631715,7592464,12107147 ,12850135 20817675</pubmed> | |
| − | <pubmed>8631715,7592464,12107147 ,12850135 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:29, 18 June 2013
- Description: menaquinol:cytochrome c oxidoreductase (cytochrome b subunit), component of the cytochrome bc complex
| Gene name | qcrB |
| Synonyms | bfcB, petB |
| Essential | no |
| Product | menaquinol:cytochrome c oxidoreductase (cytochrome b subunit) |
| Function | respiration |
| Gene expression levels in SubtiExpress: qcrB | |
| MW, pI | 25 kDa, 7.441 |
| Gene length, protein length | 672 bp, 224 aa |
| Immediate neighbours | qcrC, qcrA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 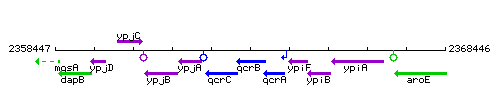
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed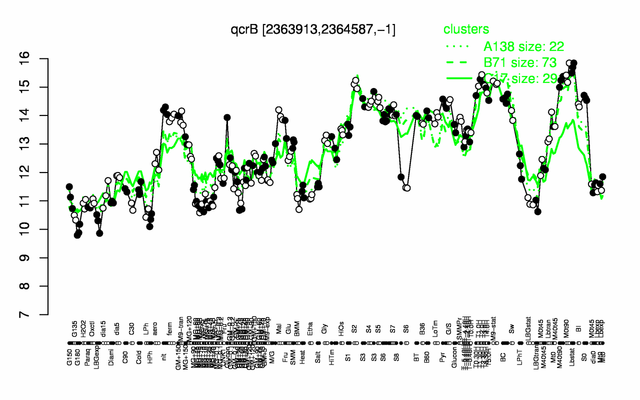
| |
Contents
Categories containing this gene/protein
respiration, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22550
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase M20A family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P46912
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
G Sun, E Sharkova, R Chesnut, S Birkey, M F Duggan, A Sorokin, P Pujic, S D Ehrlich, F M Hulett
Regulators of aerobic and anaerobic respiration in Bacillus subtilis.
J Bacteriol: 1996, 178(5);1374-85
[PubMed:8631715]
[WorldCat.org]
[DOI]
(P p)
J Yu, L Hederstedt, P J Piggot
The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization.
J Bacteriol: 1995, 177(23);6751-60
[PubMed:7592464]
[WorldCat.org]
[DOI]
(P p)