Difference between revisions of "PyrB"
| Line 57: | Line 57: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 89: | Line 87: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId= | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3R7D 3R7D] {{PubMed|21663747}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P05654 P05654] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P05654 P05654] | ||
| Line 134: | Line 132: | ||
=References= | =References= | ||
| − | <pubmed>7868607,8206849,12896995,3015959,7840626,6427186,1709162,3937019,17218307, 17322189 </pubmed> | + | <pubmed>7868607,8206849,12896995,3015959,7840626,6427186,1709162,3937019,17218307, 17322189 21663747 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:54, 15 June 2011
- Description: aspartate carbamoyltransferase
| Gene name | pyrB |
| Synonyms | |
| Essential | no |
| Product | aspartate carbamoyltransferase |
| Function | pyrimidine biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Pyrimidines, Nucleotides (regulation) | |
| MW, pI | 34 kDa, 5.341 |
| Gene length, protein length | 912 bp, 304 aa |
| Immediate neighbours | pyrP, pyrC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 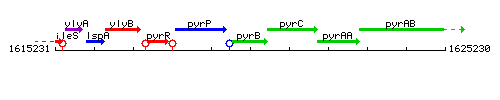
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Carbamoyl phosphate + L-aspartate = phosphate + N-carbamoyl-L-aspartate (according to Swiss-Prot)
- Protein family: ATCase/OTCase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-303 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: P05654
- KEGG entry: [3]
- E.C. number: 2.1.3.2
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulatory mechanism:
- PyrR: RNA switch, transcription termination/ antitermination (in the presence of uridine nucleotides: termination, in their absence: antitermination) PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References