Difference between revisions of "PycA"
(→References) |
(→Expression and regulation) |
||
| Line 98: | Line 98: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** subject to positive stringent control upon lysine starvation {{PubMed|20081037}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** stringent response: due to presence of adenines at +1 and +2 positions of the transcript {{PubMed|20081037}} | ||
* '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | ||
Revision as of 17:10, 19 January 2010
- Description: pyruvate carboxylase
| Gene name | pycA |
| Synonyms | ylaP |
| Essential | no |
| Product | pyruvate carboxylase |
| Function | replenishment of the oxaloacetate pool |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 127 kDa, 5.407 |
| Gene length, protein length | 3444 bp, 1148 aa |
| Immediate neighbours | ftsW, ctaA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 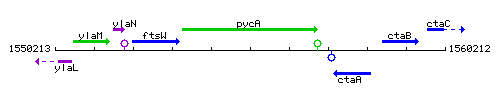
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU14860
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): biotin
- Effectors of protein activity:
- Interactions:
- Localization: membrane associated PubMed
Database entries
- Structure: 3BG5 (S. aureus)
- UniProt: Q9KWU4
- KEGG entry: [3]
- E.C. number: 6.4.1.1
Additional information
PycA binds to StrepTactin, and may be co-purified when purifying Strep-tagged proteins by SPINE.
PycA is subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- subject to positive stringent control upon lysine starvation PubMed
- Regulatory mechanism:
- stringent response: due to presence of adenines at +1 and +2 positions of the transcript PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Shigeo Tojo, Kanako Kumamoto, Kazutake Hirooka, Yasutaro Fujita
Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis.
J Bacteriol: 2010, 192(6);1573-85
[PubMed:20081037]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)