Difference between revisions of "PurD"
| Line 55: | Line 55: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU06530&redirect=T BSU06530] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/purEKBCSQLFMNHD.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/purEKBCSQLFMNHD.html] | ||
| Line 89: | Line 90: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU06530&redirect=T BSU06530] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2XCL 2XCL] | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2XCL 2XCL] | ||
Revision as of 13:08, 2 April 2014
- Description: phosphoribosylglycinamide synthetase
| Gene name | purD |
| Synonyms | |
| Essential | no |
| Product | phosphoribosylglycinamide synthetase |
| Function | purine biosynthesis |
| Gene expression levels in SubtiExpress: purD | |
| Metabolic function and regulation of this protein in SubtiPathways: purD | |
| MW, pI | 45 kDa, 4.604 |
| Gene length, protein length | 1266 bp, 422 aa |
| Immediate neighbours | purH, yezC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed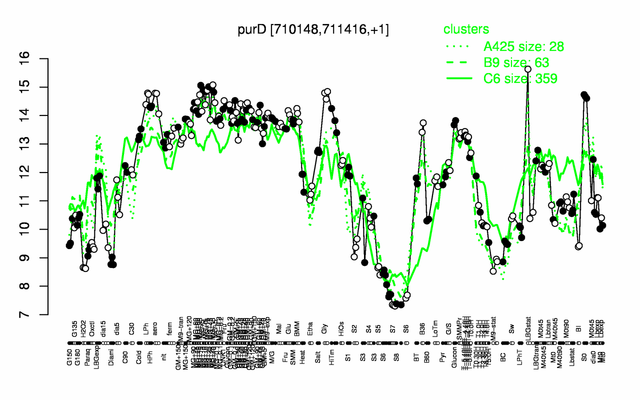
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06530
Phenotypes of a mutant
Database entries
- BsubCyc: BSU06530
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + 5-phospho-D-ribosylamine + glycine = ADP + phosphate + N(1)-(5-phospho-D-ribosyl)glycinamide (according to Swiss-Prot)
- Protein family: ATP-grasp domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU06530
- Structure: 2XCL
- UniProt: P12039
- KEGG entry: [3]
- E.C. number: 6.3.4.13
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
M Weng, P L Nagy, H Zalkin
Identification of the Bacillus subtilis pur operon repressor.
Proc Natl Acad Sci U S A: 1995, 92(16);7455-9
[PubMed:7638212]
[WorldCat.org]
[DOI]
(P p)
D J Ebbole, H Zalkin
Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis.
J Biol Chem: 1987, 262(17);8274-87
[PubMed:3036807]
[WorldCat.org]
(P p)